R.version4 Managing Packages and Workflows
4.1 Introduction
In Lab 4, we will explore essential practices that will enhance your efficiency and effectiveness as an R programmer. You will discover how to extend R’s capabilities by installing and loading packages, how to ensure that your analyses are reproducible by using RStudio Projects, and how to proficiently import and export datasets in various formats. These skills are essential for any data analyst or data scientist, as they enable you to work with a wide range of data sources, maintain the integrity of your analyses, and share your work with others in a consistent and reliable manner.
4.2 Learning Objectives
By the end of this lab, you will be able to:
Install and Load Packages in R
Learn how to find, install, and load packages from CRAN and other repositories, thereby extending the functionality of R for your data analysis tasks.Ensure Reproducibility with R and RStudio Projects
Set up and manage RStudio Projects to organise your work, understand the concept of the working directory, and adopt best practices to make your data analyses reproducible and shareable.Import and Export Datasets in Various Formats
Import data into R from different file types, such as CSV, Excel, and SPSS, using appropriate packages and functions. Export your data frames and analysis results to various formats for sharing or reporting.
By completing this lab, you will enhance your ability to manage and analyse data in R more efficiently. You will also ensure that your work is organised, reproducible, and ready to share with others. These foundational skills will support your development as a proficient R programmer and data analyst.
4.3 Prerequisites
Before starting this lab, you should have:
Completed Lab 3 or have a basic understanding of writing custom functions in R.
Familiarity with R’s data structures and basic data manipulation.
An interest in organizing, documenting, and sharing analytical workflows efficiently.
4.4 Understanding Packages and Libraries in R
In R, a package is a collection of functions, data, and code that extends the basic functionality of R. Think of it as a specialised toolkit for particular tasks or topics. For example, packages like tidyr and janitor facilitate data wrangling, while others focus on graphics, modelling, or data import and export.
A library is a location on your computer’s file system where installed packages are stored. When you install a package, it is saved in a library so that you can easily access it in future R sessions.
4.5 Compiling R Packages from Source
You may occasionally need additional tools to compile R packages from source, depending on your operating system:
-
Windows
Windows does not support code compilation natively. Therefore, you need Rtools, which provides the necessary software, including compilers and libraries, to build R packages from source. You can download Rtools from CRAN: https://cran.rstudio.com/bin/windows/Rtools/. After installing the appropriate version of Rtools, R will automatically detect it.
NoteTo check your R version, run the following code in your console:
To verify that Rtools is correctly installed, you can run the following code in your console:
Sys.which("make") -
Mac OS
On macOS, you need the Xcode Command Line Tools, which provide similar capabilities to Rtools on Windows. You can install Xcode from the Mac App Store:
http://itunes.apple.com/us/app/xcode/id497799835?mt=12 or install the Command Line Tools directly by running:
xcode-select --install -
Linux
Most Linux distributions already come with the necessary tools for compiling packages. If additional developer tools are needed, you can install them via your package manager, usually by installing packages like
build-essentialor similar for your Linux distribution.NoteOn Debian/Ubuntu, you can install the essential software for R package development and LaTeX (if needed for documentation) with:
sudo apt-get install r-base-dev texlive-fullTo ensure all dependencies for building R itself from source are met, you can run:
sudo apt-get build-dep r-base-core
4.6 Experiment 4.1: Installing and Loading Packages
As you progress in R, you will frequently need functions that are not included in the base R installation. These are provided by packages, which you can easily install and load into your R environment.
4.6.1 Installing Packages from CRAN
The Comprehensive R Archive Network (CRAN) hosts thousands of packages. To install a package from CRAN, use:
install.packages("package_name")Replace package_name with the name of the package you want to install.
For example, to install the tidyverse package, use:
install.packages("tidyverse")Similarly, to install the janitor package, use:
install.packages("janitor")Remember to enclose the package name in quotes—either double ("package_name") or single ('package_name').
4.6.2 Installing Packages from External Repositories
Some packages may not be available on CRAN but can be installed from GitHub or GitLab. First, install a helper package such as devtools or remotes:
install.packages("devtools")
# or
install.packages("remotes")Then, to install a package from GitHub, for example openintro package, use:
devtools::install_github("OpenIntroStat/openintro")
# or
remotes::install_github("OpenIntroStat/openintro")You can also install development versions of packages using these helper packages. For instance:
#|
remotes::install_github("datalorax/equatiomatic")4.6.3 Loading Packages
Once a package has been installed, you need to load it into your R session to use its functions. You can do this by calling the library() function, as demonstrated in the code cell below:
library(package_name)Here, package_name refers to the specific package you want to load into the R environment. For example, to load the tidyverse package:
#> ── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
#> ✔ dplyr 1.1.4 ✔ readr 2.1.5
#> ✔ forcats 1.0.0 ✔ stringr 1.5.1
#> ✔ ggplot2 3.5.1 ✔ tibble 3.2.1
#> ✔ lubridate 1.9.3 ✔ tidyr 1.3.1
#> ✔ purrr 1.0.2
#> ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
#> ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsThis command loads core tidyverse packages essential for most data analysis project1. Other installed packages can also be loaded in such manner:
If you run this code and get the error message there is no package called "bulkreadr", you’ll need to first install it, then run library() once again.
install.packages("bulkreadr")
library(bulkreadr)4.6.4 Using Functions from a Package
When working with R packages, there are two primary ways to use a function from a package:
- Load the Package and Call the Function Directly
You can load the package into your R session using the library() function, and then call the desired function by its name. For example:
#|
# Load the janitor package
library(janitor)
# Use the clean_names() function
clean_names(iris)#> # A tibble: 150 × 5
#> sepal_length sepal_width petal_length petal_width species
#> <dbl> <dbl> <dbl> <dbl> <fct>
#> 1 5.1 3.5 1.4 0.2 setosa
#> 2 4.9 3 1.4 0.2 setosa
#> 3 4.7 3.2 1.3 0.2 setosa
#> 4 4.6 3.1 1.5 0.2 setosa
#> 5 5 3.6 1.4 0.2 setosa
#> 6 5.4 3.9 1.7 0.4 setosa
#> 7 4.6 3.4 1.4 0.3 setosa
#> 8 5 3.4 1.5 0.2 setosa
#> 9 4.4 2.9 1.4 0.2 setosa
#> 10 4.9 3.1 1.5 0.1 setosa
#> # ℹ 140 more rowsIn this approach, you only need to load the package once in your session, and then you can use its functions without specifying the package name.
- Use the
::Operator to Call the Function Without Loading the Package
The :: operator is used to call a function from a specific package without loading the entire package into your R session. This is particularly useful when:
Avoiding Namespace Conflicts: If multiple packages have functions with the same name, :: ensures you use the correct one.
Improving Code Clarity: It makes your code more readable by clearly indicating which package a function comes from.
Reducing Memory Usage: Only the specific function is accessed, not the entire package.
The syntax is:
packageName::functionName(arguments)where:
packageName: The name of the package where the function resides.functionName: The specific function you want to use from the package.
Example
# Using the double colon to access clean_names() from janitor package
janitor::clean_names(iris)#> # A tibble: 150 × 5
#> sepal_length sepal_width petal_length petal_width species
#> <dbl> <dbl> <dbl> <dbl> <fct>
#> 1 5.1 3.5 1.4 0.2 setosa
#> 2 4.9 3 1.4 0.2 setosa
#> 3 4.7 3.2 1.3 0.2 setosa
#> 4 4.6 3.1 1.5 0.2 setosa
#> 5 5 3.6 1.4 0.2 setosa
#> 6 5.4 3.9 1.7 0.4 setosa
#> 7 4.6 3.4 1.4 0.3 setosa
#> 8 5 3.4 1.5 0.2 setosa
#> 9 4.4 2.9 1.4 0.2 setosa
#> 10 4.9 3.1 1.5 0.1 setosa
#> # ℹ 140 more rowsThe clean_names() function, in this case, returns the iris data frame with column names formatted in a consistent and clean style.
Ensure that the janitor package is installed before using its functions. If it’s not installed, you can install it using install.packages(“janitor”).
As shown in Table 4.1, using library() attaches the entire package, while the :: operator allows you to call specific functions without loading the entire package. This difference can significantly impact memory usage and clarity in your scripts, especially when you’re only using a few functions from a large package.
library() and :: Operator
| Aspect | library() |
:: Operator |
|---|---|---|
| Loading Behavior | Attaches the entire package to the session | Accesses specific functions without loading the package |
| Namespace Conflicts | Potential for conflicts if multiple packages have functions with the same name | Avoids conflicts by specifying the package |
| Memory Usage | Loads all exported objects, increasing memory usage | Minimal memory usage as only specific functions are accessed |
| Code Verbosity | Less verbose; functions can be called directly | More verbose; requires prefixing with package name |
| Use Cases | When using multiple functions from a package extensively | When using only a few functions or to avoid conflicts |
4.6.5 Practice Quiz 4.1
Question 1:
Imagine that you want to install the shiny package from CRAN. Which command should you use?
install.packages("shiny")-
library("shiny")
-
install.packages(shiny)
require("shiny")
Question 2:
What must you do after installing a package before you can use it in your current session?
- Restart R
- Run
install.packages()again
- Load it with
library()
- Convert the package into a dataset
Question 3:
If you want to install a package that is not on CRAN (e.g., from GitHub), which additional package would be helpful?
-
installer
-
rio
-
devtools
github_install
Question 4:
Which function would you use to update all outdated packages in your R environment?
Question 5:
Which function can be used to check the version of an installed package?
-
version()
-
packageVersion()
-
libraryVersion()
install.packages()
4.7 Experiment 4.2: Ensuring Reproducibility with R and RStudio Projects
Reproducibility is vital in data science. It allows analyses to be revisited, results to be verified, and insights to be shared seamlessly, whether with collaborators or for future reference. With R and RStudio Projects, you can create a self-contained workspace that keeps your work organised and reproducible.
4.7.1 Working Directory and Paths
Your working directory is where R looks for files to read and where it saves outputs. You can find your current working directory in two ways:
-
In the Console: At the top of the RStudio console, the current working directory is displayed.
-
Using Code: Run the following command:
getwd()[1] "C:/Users/Ezekiel Adebayo/Desktop/stock-market"If you’re not using an RStudio project, you’ll need to set the working directory manually each time. For instance:
setwd("/path/to/your/data_analysis")TipYou can use the keyboard shortcut
Ctrl + Shift + Hin RStudio to quickly choose your working directory.
Absolute Paths: Start from the root of your file system (e.g.,
C:/Users/YourName/Documents/data.csv). These paths are specific to your computer and should be avoided in shared scripts.Relative Paths: Refer to files relative to your working directory (e.g.,
data/data.csv). Using relative paths ensures portability and makes your scripts easier to share and reuse.
4.7.2 RStudio Projects
RStudio Projects provide a centralised environment for all your analyses: data files, scripts, figures, outputs, and documentation. This setup ensures your work remains organised, consistent, and reproducible.
Benefits of RStudio Projects
Organisation
All project files are stored in one place, making it easy to navigate and manage.Relative Paths
RStudio automatically sets the working directory to the project folder, allowing you to use relative paths (e.g.,data/my_data.csv). This eliminates the need for hardcoding absolute paths, ensuring your code works on any system.Consistency Across Systems
When you open your RStudio project on another computer, it recognises the same folder structure, making your work portable and adaptable.Reproducibility
With all components centralised and paths consistent, RStudio Projects enable others to replicate your workflow effortlessly. Everything needed to reproduce your analysis is neatly packaged and ready to go.
4.7.3 How RStudio Projects Organize Your Work
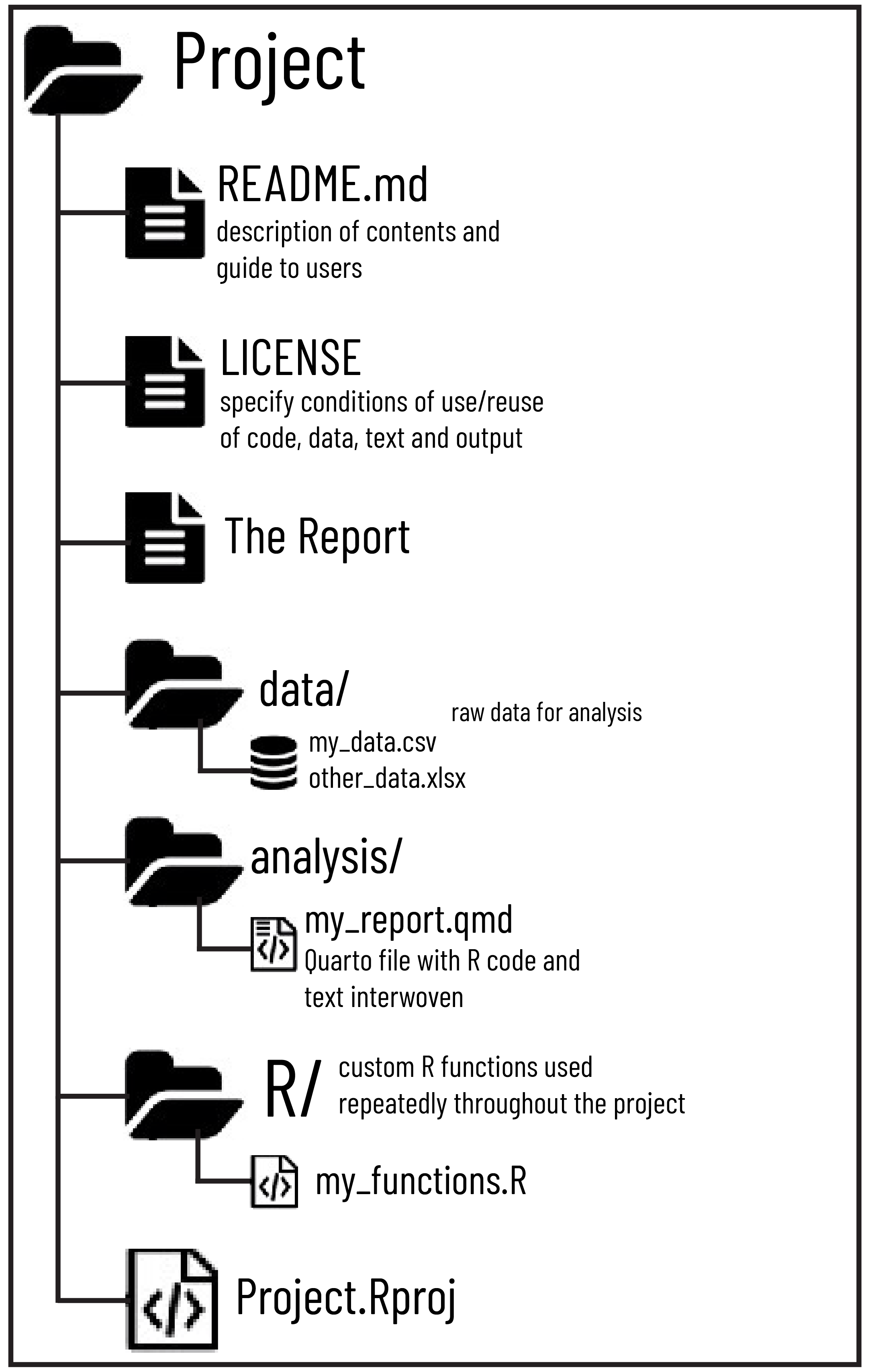
When you set up an RStudio Project, it acts as the root folder for your analysis. A recommended structure is shown in Figure 4.2 below:
Top-Level Files
Files likeREADME.mdprovide an overview of the project, while aLICENSEfile outlines terms of use for the code and data. These files are essential for collaboration and transparency.Data Folder (
data/)
This folder contains raw data files in formats like.csvor.xlsx. Raw data is stored here in its original state to preserve reproducibility.Analysis Folder (
analysis/)
Scripts or dynamic documents, such as Quarto files (.qmd) or R Markdown files (.Rmd), go here. These documents combine R code, narrative text, and visualizations, serving as the heart of your analysis.Custom Functions Folder (
R/)
Reusable R scripts containing custom functions are saved here. These can be sourced into analysis scripts to keep your workflow modular and efficient.
This structure, combined with relative paths, ensures a clean, logical, and reproducible workflow.
4.7.4 Setting Up Your RStudio Project
Let’s create a new RStudio project. You can do this by following these simple steps:
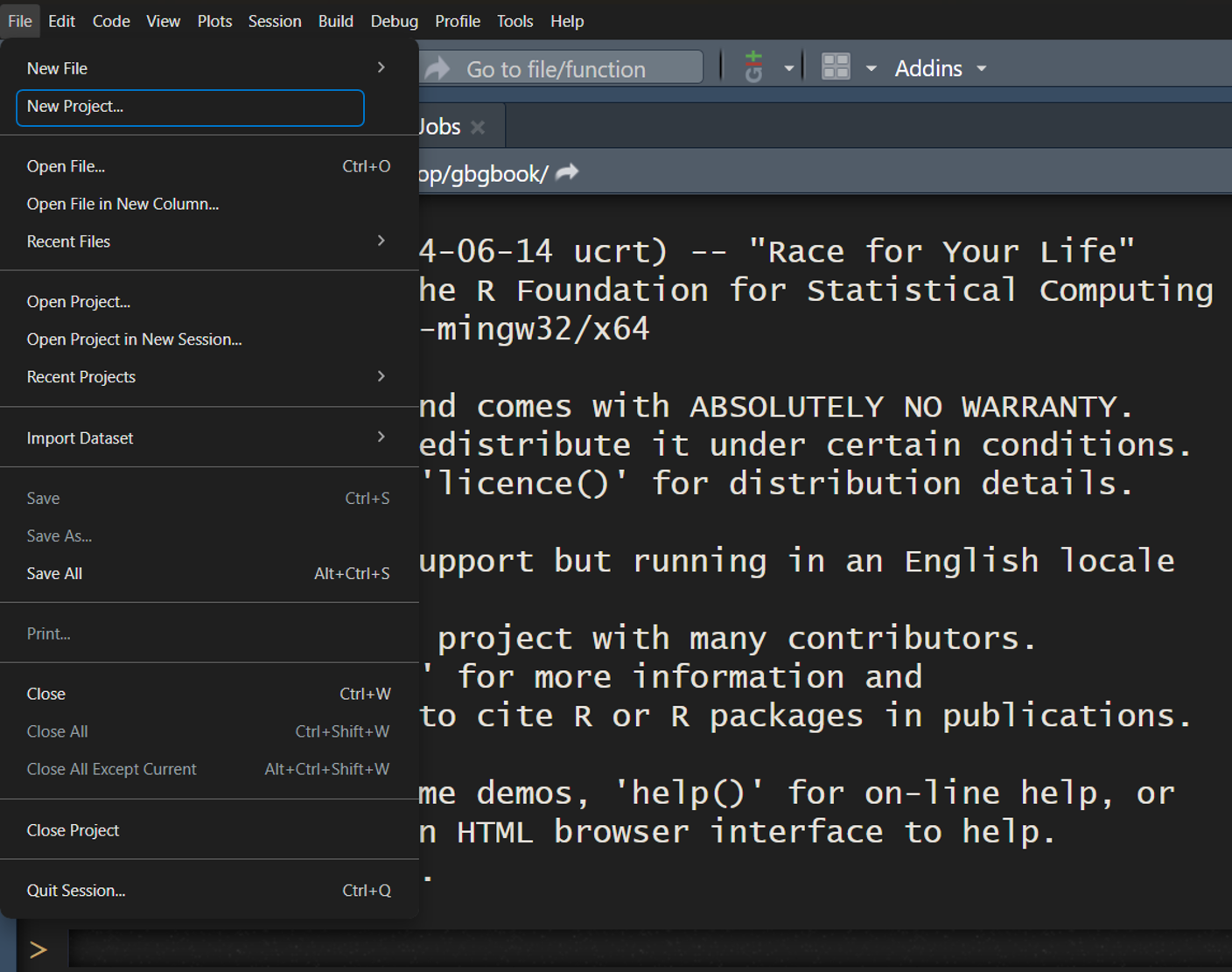
4.7.4.1 Step 1: Create a New Project
-
Go to:
File → New Projectin RStudio
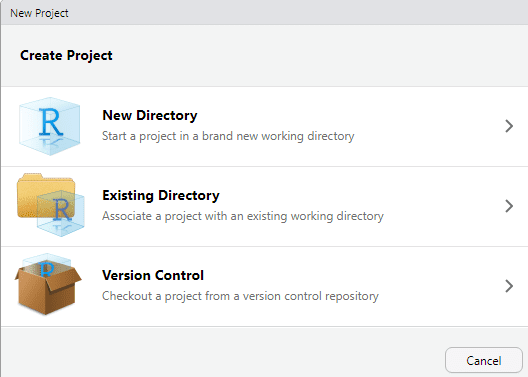
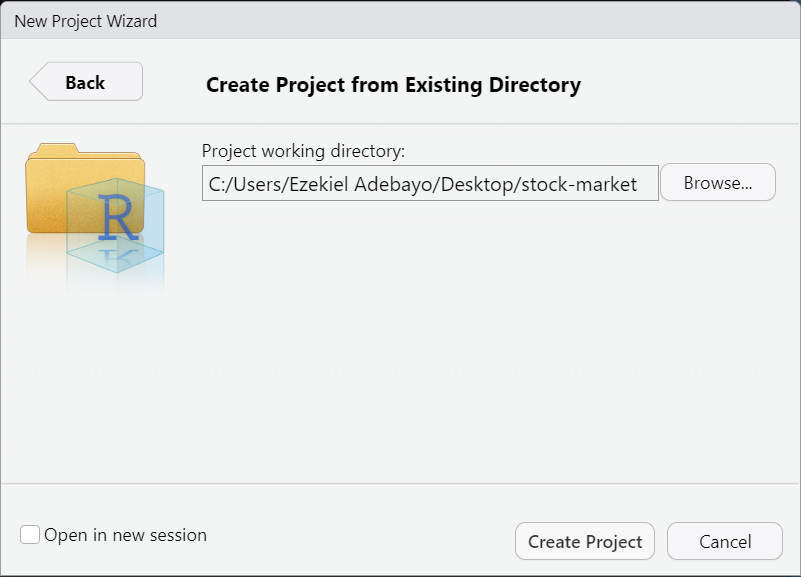
-
Choose:
Existing Directory
Select the folder you want to use as your project’s working directory and RStudio will create a project file (
.Rproj).Click:
Create Project
Once you click “Create Project”, you’re all set! You’ll be inside your new RStudio project.
4.7.4.2 Step 2: Arrange Your Files
Organise your project files into the following structure:
data/: Store raw data files here in formats such as.csvor.xlsx.analysis/: Place your analysis scripts or reports in this folder. For example, you might use a Quarto file (my_report.qmd) to combine R code, narrative text, and visualisations.R/: Save reusable R functions in this folder, and source them into your scripts as needed.
4.7.4.3 Step 3: Use Relative Paths
Always refer to files using paths relative to your project folder. For example:
data <- read.csv("data/my_data.csv")4.7.4.4 Step 4: Add Documentation
Include a README.md file to document the purpose, structure, and usage of your project. Add a LICENSE file to define terms of use.
From now on, whenever you open this project (by clicking the .Rproj file), RStudio will automatically set your working directory, allowing you to use relative paths easily as shown in Figure 4.7
4.7.5 Practice Quiz 4.2
Question 1:
What is a key advantage of using RStudio Projects?
- They automatically install packages.
- They allow you to use absolute paths easily.
- They set the working directory to the project folder, enabling relative paths.
- They prevent package updates.
Question 2:
Which file extension identifies an RStudio Project file?
-
.Rdata
-
.Rproj
-
.Rmd
.Rscript
Question 3:
Why are relative paths preferable in a collaborative environment?
- They are shorter and easier to type.
- They change automatically when you move files.
- They ensure that the code works regardless of the user’s file system structure.
- They are required for Git version control.
4.8 Experiment 4.3: Importing and Exporting Data in R
Data import and export are vital steps in data science. With R, you can load data from spreadsheets, databases, and many other formats, and subsequently save your processed results. Figure 4.8 below illustrates some popular R packages for data import:
Packages like readr, readxl, and haven are part of the tidyverse, and therefore are pre-installed when you install the tidyverse. You do not need to install these packages individually. For a complete list of tidyverse packages, see the following code:
tidyverse::tidyverse_packages()#> [1] "broom" "conflicted" "cli" "dbplyr"
#> [5] "dplyr" "dtplyr" "forcats" "ggplot2"
#> [9] "googledrive" "googlesheets4" "haven" "hms"
#> [13] "httr" "jsonlite" "lubridate" "magrittr"
#> [17] "modelr" "pillar" "purrr" "ragg"
#> [21] "readr" "readxl" "reprex" "rlang"
#> [25] "rstudioapi" "rvest" "stringr" "tibble"
#> [29] "tidyr" "xml2" "tidyverse"You don’t need to install any of these packages individually since they’re all included with the tidyverse installation.
R offers some excellent packages that simplify the processes of importing and exporting data. In the sections below, we will explore a few commonly used packages and functions that are essential when working with data in R.
4.8.1 Flat Files
Flat files, such as CSV files, are among the most common formats for data storage and exchange. The readr package (a core component of the tidyverse) provides functions specifically designed to handle flat files. The two primary functions are:
read_csv(): This function imports data from a CSV file into R as a data frame, akin to loading data directly from a spreadsheet.write_csv(): Once your data analysis is complete, this function exports your data frame to a CSV file. It is particularly useful for sharing your results or maintaining a backup.
Example 1: Reading CSV Data From a File
Suppose you have a file named cleveland-heart-disease-database.csv located in the r-data folder. You can import this flat file into R as follows:
library(tidyverse)
heart_disease_data <- read_csv("r-data/cleveland-heart-disease-database.csv")
heart_disease_data#> # A tibble: 303 × 14
#> age sex `chest pain type` resting blood pressur…¹ serum cholestoral in…²
#> <dbl> <chr> <chr> <dbl> <dbl>
#> 1 63 male typical angina 145 233
#> 2 67 male asymptomatic 160 286
#> 3 67 male asymptomatic 120 229
#> 4 37 male non-anginal pain 130 250
#> 5 41 female atypical angina 130 204
#> 6 56 male atypical angina 120 236
#> 7 62 female asymptomatic 140 268
#> 8 57 female asymptomatic 120 354
#> 9 63 male asymptomatic 130 254
#> 10 53 male asymptomatic 140 203
#> # ℹ 293 more rows
#> # ℹ abbreviated names: ¹`resting blood pressure in mm Hg`,
#> # ²`serum cholestoral in mg/dl`
#> # ℹ 9 more variables: `fasting blood sugar > 120 mg/dl` <lgl>,
#> # `resting electrocardiographic results` <dbl>,
#> # `maximum heart rate achieved` <dbl>, `exercise induced angina` <chr>,
#> # `ST depression` <dbl>, `slope of the peak exercise ST segment` <chr>, …For information on downloading the data, please refer to Appendix B.1. Should you encounter any errors when executing the code, first set up your RStudio project as described in Section 4.8.5, then re-run the code.
When you run read_csv(), you will notice a message detailing the number of rows and columns, the delimiter used, and information regarding the column types. This ensures that your data is read correctly. You may also specify how missing values are represented via the na argument. For example, setting na = "?" tells read_csv() to treat the ? symbol as NA in the dataset:
heart_disease_data <- read_csv("r-data/cleveland-heart-disease-database.csv", na = "?")
heart_disease_data#> # A tibble: 303 × 14
#> age sex `chest pain type` resting blood pressur…¹ serum cholestoral in…²
#> <dbl> <chr> <chr> <dbl> <dbl>
#> 1 63 male typical angina 145 233
#> 2 67 male asymptomatic 160 286
#> 3 67 male asymptomatic 120 229
#> 4 37 male non-anginal pain 130 250
#> 5 41 female atypical angina 130 204
#> 6 56 male atypical angina 120 236
#> 7 62 female asymptomatic 140 268
#> 8 57 female asymptomatic 120 354
#> 9 63 male asymptomatic 130 254
#> 10 53 male asymptomatic 140 203
#> # ℹ 293 more rows
#> # ℹ abbreviated names: ¹`resting blood pressure in mm Hg`,
#> # ²`serum cholestoral in mg/dl`
#> # ℹ 9 more variables: `fasting blood sugar > 120 mg/dl` <lgl>,
#> # `resting electrocardiographic results` <dbl>,
#> # `maximum heart rate achieved` <dbl>, `exercise induced angina` <chr>,
#> # `ST depression` <dbl>, `slope of the peak exercise ST segment` <chr>, …Example 2: Writing to a CSV File
After processing or analysing your data, you might wish to save your results. The write_csv() function writes the data to disk, enabling you to share your cleaned or transformed data with others. The key arguments are:
x: The data frame to be saved.file: the destination file path.
For example:
write_csv(heart_disease_data, "processed-cleveland-heart-disease-data.csv")Additional arguments allow you to control the representation of missing values (using na) and whether to append to an existing file (using append). For instance:
4.8.2 Spreadsheets
Microsoft Excel is a widely used application that organises data into worksheets within a single workbook2. The readxl package is used to import Excel spreadsheets (e.g. .xlsx files) into R, while the writexl package is used to export data frames to Excel files.
The primary functions are:
read_xlsx(): Imports an Excel file into R. You can specify the worksheet containing your data by using thesheetargument.write_xlsx(): Export your data frame to an Excel file—ideal for sharing your work with colleagues who prefer Excel.
Example 1: Reading Excel Spreadsheets
In this example, we import data from an Excel spreadsheet using the readxl package. Although readxl is not part of the core tidyverse, it is installed automatically with the tidyverse, so you must load it explicitly:
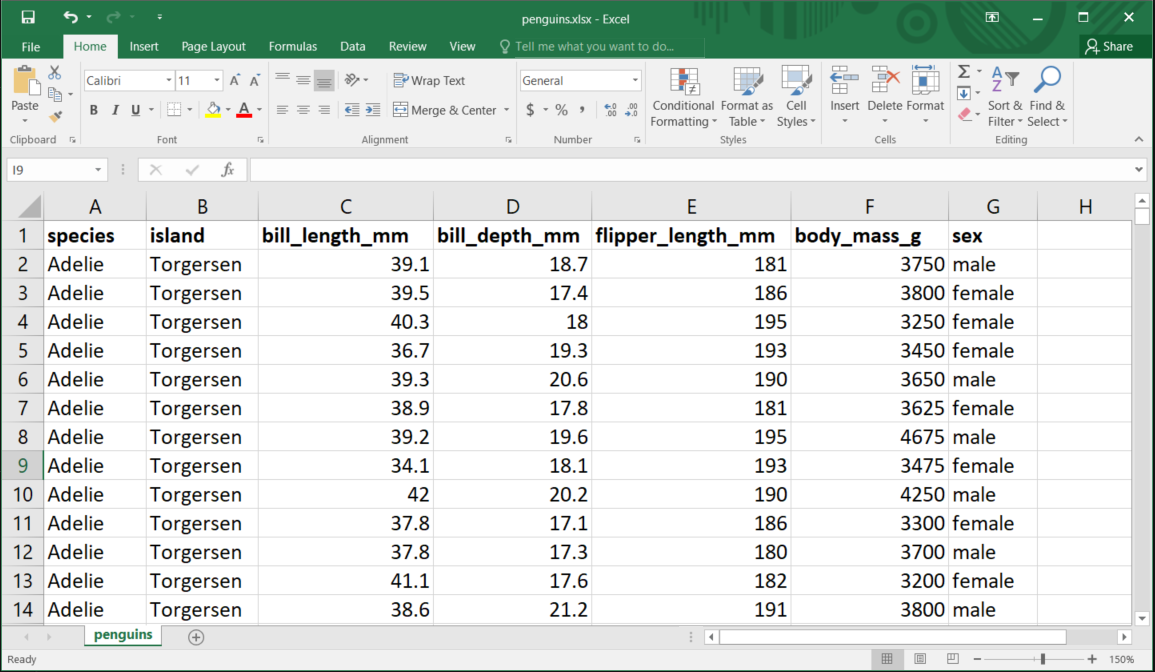
The spreadsheet, as shown in Figure 4.9, can be downloaded from https://docs.google.com/spreadsheets/d/107H-n59gDw0QoIktksU9wv6Iro4TGUAOgmQJW6pb19Y
The first argument of read_xlsx() is the file path:
penguins <- read_xlsx("r-data/penguins.xlsx")This function reads the file as a tibble3:
penguins#> # A tibble: 337 × 7
#> species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Adelie Torgersen 39.1 18.7 181 3750
#> 2 Adelie Torgersen 39.5 17.4 186 3800
#> 3 Adelie Torgersen 40.3 18 195 3250
#> 4 Adelie Torgersen 36.7 19.3 193 3450
#> 5 Adelie Torgersen 39.3 20.6 190 3650
#> 6 Adelie Torgersen 38.9 17.8 181 3625
#> 7 Adelie Torgersen 39.2 19.6 195 4675
#> 8 Adelie Torgersen 34.1 18.1 193 3475
#> 9 Adelie Torgersen 42 20.2 190 4250
#> 10 Adelie Torgersen 37.8 17.1 186 3300
#> # ℹ 327 more rows
#> # ℹ 1 more variable: sex <chr>This dataset contains data on 337 penguins and includes seven variables for each species.
Example 2: Reading Worksheets
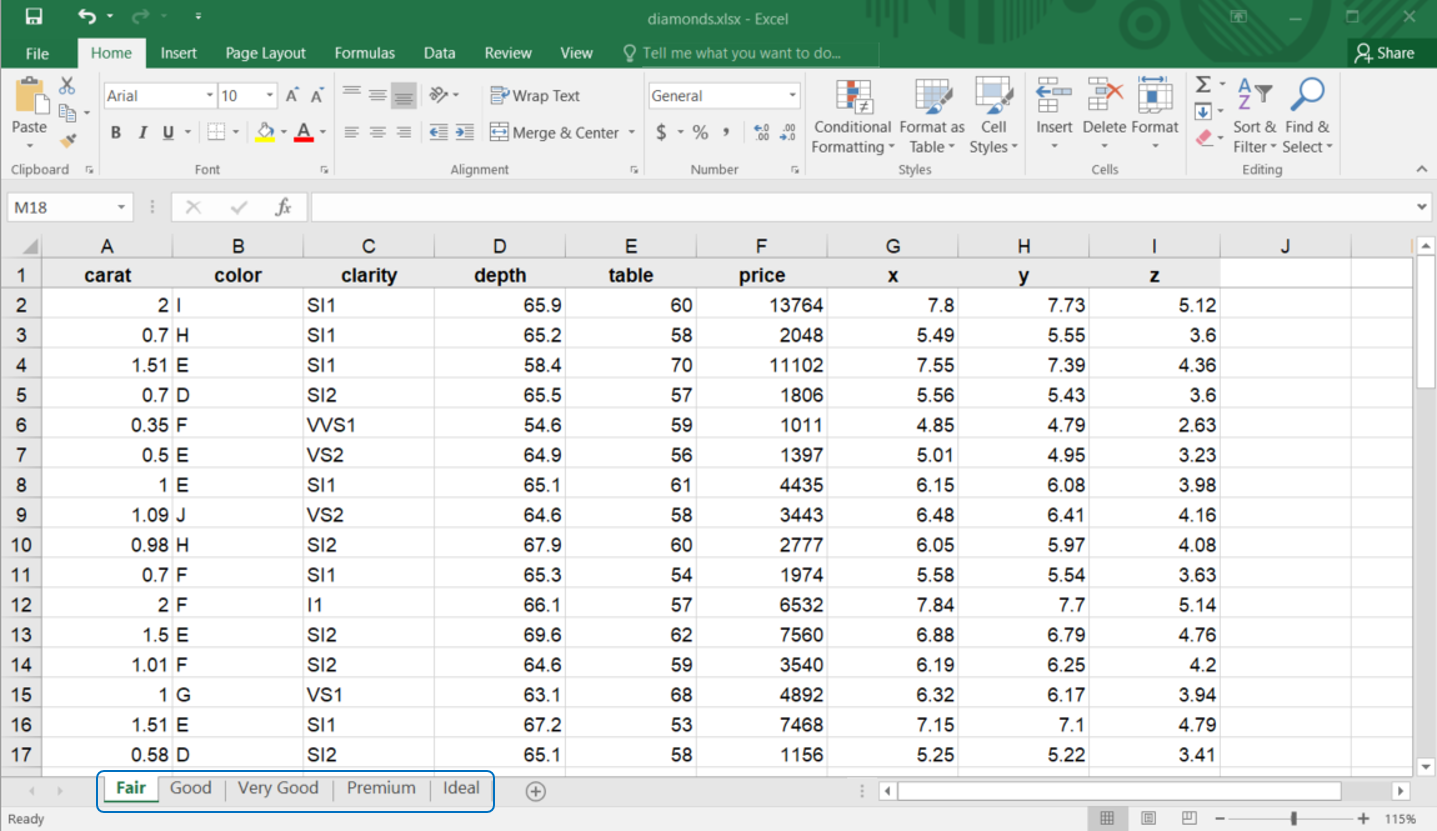
Spreadsheets may contain multiple worksheets. Figure 4.10 illustrates an Excel workbook with several sheets. The data, sourced from the ggplot2 package, is available at https://docs.google.com/spreadsheets/d/1izO0mHu3L9AMySQUXGDn9GPs1n-VwGFSEoAKGhqVQh0. Each worksheet contains information on diamond prices for different cuts.
You can read a specific worksheet by using the sheet argument in read_xlsx(). By default, the first worksheet is read.
diamonds_fair <- read_xlsx("r-data/diamonds.xlsx", sheet = "Fair")
diamonds_fair#> # A tibble: 60 × 9
#> carat color clarity depth table price x y z
#> <dbl> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 2 I SI1 65.9 60 13764 7.8 7.73 5.12
#> 2 0.7 H SI1 65.2 58 2048 5.49 5.55 3.6
#> 3 1.51 E SI1 58.4 70 11102 7.55 7.39 4.36
#> 4 0.7 D SI2 65.5 57 1806 5.56 5.43 3.6
#> 5 0.35 F VVS1 54.6 59 1011 4.85 4.79 2.63
#> 6 0.5 E VS2 64.9 56 1397 5.01 4.95 3.23
#> 7 1 E SI1 65.1 61 4435 6.15 6.08 3.98
#> 8 1.09 J VS2 64.6 58 3443 6.48 6.41 4.16
#> 9 0.98 H SI2 67.9 60 2777 6.05 5.97 4.08
#> 10 0.7 F SI1 65.3 54 1974 5.58 5.54 3.63
#> # ℹ 50 more rowsIf numerical data is read as text because the string “NA” is not automatically recognised as a missing value, you may correct this by specifying the na argument:
read_excel("r-data/diamonds.xlsx", sheet = "Fair", na = "NA")#> # A tibble: 60 × 9
#> carat color clarity depth table price x y z
#> <dbl> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 2 I SI1 65.9 60 13764 7.8 7.73 5.12
#> 2 0.7 H SI1 65.2 58 2048 5.49 5.55 3.6
#> 3 1.51 E SI1 58.4 70 11102 7.55 7.39 4.36
#> 4 0.7 D SI2 65.5 57 1806 5.56 5.43 3.6
#> 5 0.35 F VVS1 54.6 59 1011 4.85 4.79 2.63
#> 6 0.5 E VS2 64.9 56 1397 5.01 4.95 3.23
#> 7 1 E SI1 65.1 61 4435 6.15 6.08 3.98
#> 8 1.09 J VS2 64.6 58 3443 6.48 6.41 4.16
#> 9 0.98 H SI2 67.9 60 2777 6.05 5.97 4.08
#> 10 0.7 F SI1 65.3 54 1974 5.58 5.54 3.63
#> # ℹ 50 more rowsAnother approach is to use excel_sheets() to list all the worksheets in an Excel file and then import only the ones you need.
excel_sheets("r-data/diamonds.xlsx")#> [1] "Fair" "Good" "Very Good" "Premium" "Ideal"Once the worksheet names are known, they can be imported individually:
diamonds_fair <- read_excel("r-data/diamonds.xlsx", sheet = "Fair")
diamonds_good <- read_excel("r-data/diamonds.xlsx", sheet = "Good")
diamonds_very_good <- read_excel("r-data/diamonds.xlsx", sheet = "Very Good")
diamonds_premium <- read_excel("r-data/diamonds.xlsx", sheet = "Premium")
diamonds_ideal <- read_excel("r-data/diamonds.xlsx", sheet = "Ideal")In this instance, the complete diamonds dataset is distributed across five worksheets that share the same columns but differ in the number of rows. You can inspect their dimensions using:
dim(diamonds_fair)#> [1] 60 9dim(diamonds_good)#> [1] 49 9dim(diamonds_very_good)#> [1] 42 9dim(diamonds_premium)#> [1] 49 9dim(diamonds_ideal)#> [1] 60 9You can also combine the worksheets into one data frame by using bind_rows():
diamonds <- bind_rows(
diamonds_fair,
diamonds_good,
diamonds_very_good,
diamonds_premium,
diamonds_ideal
)
diamonds#> # A tibble: 260 × 9
#> carat color clarity depth table price x y z
#> <dbl> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 2 I SI1 65.9 60 13764 7.8 7.73 5.12
#> 2 0.7 H SI1 65.2 58 2048 5.49 5.55 3.6
#> 3 1.51 E SI1 58.4 70 11102 7.55 7.39 4.36
#> 4 0.7 D SI2 65.5 57 1806 5.56 5.43 3.6
#> 5 0.35 F VVS1 54.6 59 1011 4.85 4.79 2.63
#> 6 0.5 E VS2 64.9 56 1397 5.01 4.95 3.23
#> 7 1 E SI1 65.1 61 4435 6.15 6.08 3.98
#> 8 1.09 J VS2 64.6 58 3443 6.48 6.41 4.16
#> 9 0.98 H SI2 67.9 60 2777 6.05 5.97 4.08
#> 10 0.7 F SI1 65.3 54 1974 5.58 5.54 3.63
#> # ℹ 250 more rowsAn alternative method to import all worksheets from a workbook is provided by the read_excel_workbook() function from the bulkreadr package. This function reads the data from every sheet in an Excel workbook and returns a single appended data frame:
library(bulkreadr)
diamonds <- read_excel_workbook("r-data/diamonds.xlsx")
diamonds#> # A tibble: 260 × 9
#> carat color clarity depth table price x y z
#> <dbl> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 2 I SI1 65.9 60 13764 7.8 7.73 5.12
#> 2 0.7 H SI1 65.2 58 2048 5.49 5.55 3.6
#> 3 1.51 E SI1 58.4 70 11102 7.55 7.39 4.36
#> 4 0.7 D SI2 65.5 57 1806 5.56 5.43 3.6
#> 5 0.35 F VVS1 54.6 59 1011 4.85 4.79 2.63
#> 6 0.5 E VS2 64.9 56 1397 5.01 4.95 3.23
#> 7 1 E SI1 65.1 61 4435 6.15 6.08 3.98
#> 8 1.09 J VS2 64.6 58 3443 6.48 6.41 4.16
#> 9 0.98 H SI2 67.9 60 2777 6.05 5.97 4.08
#> 10 0.7 F SI1 65.3 54 1974 5.58 5.54 3.63
#> # ℹ 250 more rowsYou can also specify the .id argument in read_excel_workbook() to add an output column that identifies the source of each row (using either the sheet names or their positions):
diamonds <- bulkreadr::read_excel_workbook("r-data/diamonds.xlsx", .id = "sheet")
diamonds#> # A tibble: 260 × 10
#> sheet carat color clarity depth table price x y z
#> <chr> <dbl> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Fair 2 I SI1 65.9 60 13764 7.8 7.73 5.12
#> 2 Fair 0.7 H SI1 65.2 58 2048 5.49 5.55 3.6
#> 3 Fair 1.51 E SI1 58.4 70 11102 7.55 7.39 4.36
#> 4 Fair 0.7 D SI2 65.5 57 1806 5.56 5.43 3.6
#> 5 Fair 0.35 F VVS1 54.6 59 1011 4.85 4.79 2.63
#> 6 Fair 0.5 E VS2 64.9 56 1397 5.01 4.95 3.23
#> 7 Fair 1 E SI1 65.1 61 4435 6.15 6.08 3.98
#> 8 Fair 1.09 J VS2 64.6 58 3443 6.48 6.41 4.16
#> 9 Fair 0.98 H SI2 67.9 60 2777 6.05 5.97 4.08
#> 10 Fair 0.7 F SI1 65.3 54 1974 5.58 5.54 3.63
#> # ℹ 250 more rowsExample 3: Writing Data to an Excel File
After processing your Excel data in R, you may wish to export the cleaned data. The write_xlsx() function from the writexl package enables you to save your data frame as a new Excel file:
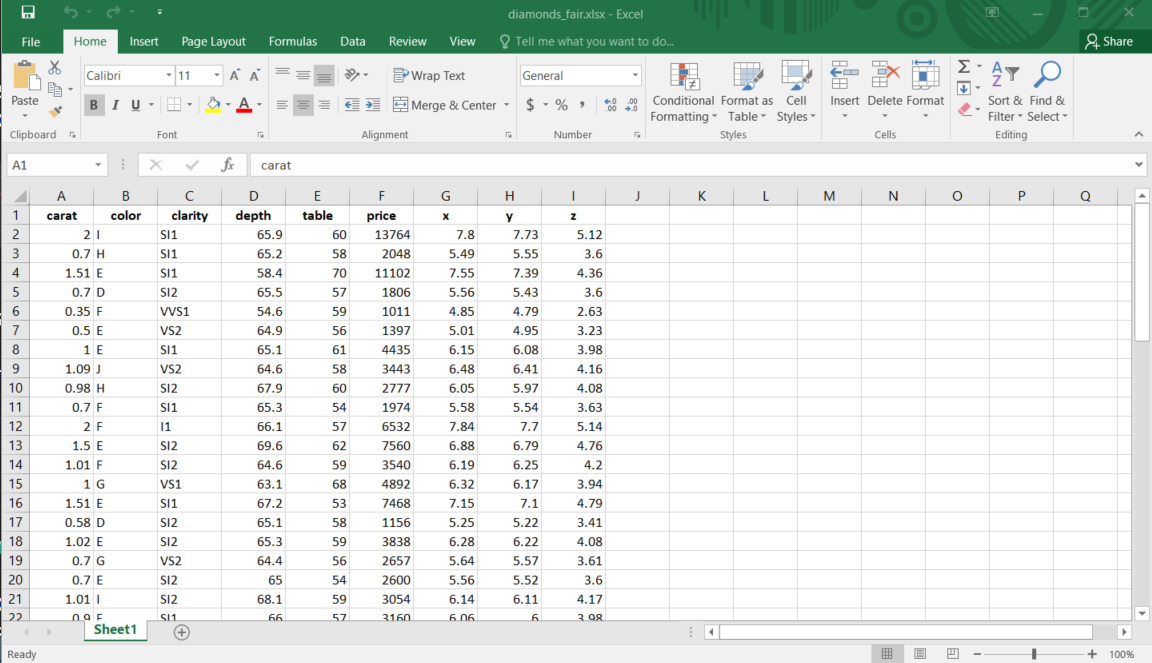
write_xlsx(diamonds_fair, path = "r-data/diamonds_fair.xlsx")Figure 4.11 shows the resulting Excel file. By default, column names are included and bolded. You can disable these features by setting the col_names and format_headers arguments to FALSE.
4.8.3 Labelled Data
Labelled data commonly originates from specialised statistical software such as SPSS, Stata, or SAS. These datasets often include value labels that describe the meaning of each code. R can import these files using the haven package. Like the readxl, haven is not part of the core tidyverse, but is installed automatically with it; however, you must load it explicitly.
For example:
-
SPSS
read_sav(): This function imports data from SPSS files (files with.savextension) into R.write_sav(): Exports a data frame from R back to SPSS format.
-
Stata
read_dta(): For Stata users, this function imports Stata files into R.write_dta(): Similarly, this function lets you export data frames to Stata format.
-
SAS
read_sas()reads .sas7bdat + .sas7bcat files andread_xpt()reads SAS transport files (versions 5 and 8).write_xpt()writes SAS transport files (versions 5 and 8).
Example 1: Reading an SPSS File
Suppose you have an SPSS file named wages.sav. You can import it using haven as follows:
#> # A tibble: 400 × 9
#> id educ south sex exper wage occup marr ed
#> <dbl> <dbl> <dbl+lbl> <dbl+l> <dbl> <dbl> <dbl+l> <dbl+l> <dbl+l>
#> 1 3 12 0 [does not live in … 0 [Mal… 17 7.5 6 [Oth… 1 [Mar… 2 [Hig…
#> 2 4 13 0 [does not live in … 0 [Mal… 9 13.1 6 [Oth… 0 [Not… 3 [Som…
#> 3 5 10 1 [lives in South] 0 [Mal… 27 4.45 6 [Oth… 0 [Not… 1 [Les…
#> 4 12 9 1 [lives in South] 0 [Mal… 30 6.25 6 [Oth… 0 [Not… 1 [Les…
#> 5 13 9 1 [lives in South] 0 [Mal… 29 20.0 6 [Oth… 1 [Mar… 1 [Les…
#> 6 14 12 0 [does not live in … 0 [Mal… 37 7.3 6 [Oth… 1 [Mar… 2 [Hig…
#> 7 17 11 0 [does not live in … 0 [Mal… 16 3.65 6 [Oth… 0 [Not… 1 [Les…
#> 8 20 12 0 [does not live in … 0 [Mal… 9 3.75 6 [Oth… 0 [Not… 2 [Hig…
#> 9 21 11 1 [lives in South] 0 [Mal… 14 4.5 6 [Oth… 1 [Mar… 1 [Les…
#> 10 23 6 1 [lives in South] 0 [Mal… 45 5.75 6 [Oth… 1 [Mar… 1 [Les…
#> # ℹ 390 more rowsIn some cases, haven’s read_sav() may not fully process labelled variables. In such instances, you can use the bulkreadr package for a more seamless import:
library(bulkreadr)
# Import SPSS data with automatic label conversion
wages_data <- read_spss_data("r-data/wages.sav")
wages_data#> # A tibble: 400 × 9
#> id educ south sex exper wage occup marr ed
#> <dbl> <dbl> <fct> <fct> <dbl> <dbl> <fct> <fct> <fct>
#> 1 3 12 does not live in South Male 17 7.5 Other Married High …
#> 2 4 13 does not live in South Male 9 13.1 Other Not married Some …
#> 3 5 10 lives in South Male 27 4.45 Other Not married Less …
#> 4 12 9 lives in South Male 30 6.25 Other Not married Less …
#> 5 13 9 lives in South Male 29 20.0 Other Married Less …
#> 6 14 12 does not live in South Male 37 7.3 Other Married High …
#> 7 17 11 does not live in South Male 16 3.65 Other Not married Less …
#> 8 20 12 does not live in South Male 9 3.75 Other Not married High …
#> 9 21 11 lives in South Male 14 4.5 Other Married Less …
#> 10 23 6 lives in South Male 45 5.75 Other Married Less …
#> # ℹ 390 more rowsExample 2: Writing to a SPSS File
After performing analyses or modifications on your SPSS data, you may wish to export the results back to an SPSS file:
wages_data |> write_sav("wages.sav")Example 3: Reading Stata File
Similarly, if you have a Stata dataset (for example, automobile.dta), you can import it using bulkreadr:
# Import Stata data with bulkreadr
automobile <- read_stata_data("r-data/automobile.dta")
glimpse(automobile)#> Rows: 32
#> Columns: 11
#> $ mpg <dbl> 21.0, 21.0, 22.8, 21.4, 18.7, 18.1, 14.3, 24.4, 22.8, 19.2, 17.8,…
#> $ cyl <dbl> 6, 6, 4, 6, 8, 6, 8, 4, 4, 6, 6, 8, 8, 8, 8, 8, 8, 4, 4, 4, 4, 8,…
#> $ disp <dbl> 160.0, 160.0, 108.0, 258.0, 360.0, 225.0, 360.0, 146.7, 140.8, 16…
#> $ hp <dbl> 110, 110, 93, 110, 175, 105, 245, 62, 95, 123, 123, 180, 180, 180…
#> $ drat <dbl> 3.90, 3.90, 3.85, 3.08, 3.15, 2.76, 3.21, 3.69, 3.92, 3.92, 3.92,…
#> $ wt <dbl> 2.620, 2.875, 2.320, 3.215, 3.440, 3.460, 3.570, 3.190, 3.150, 3.…
#> $ qsec <dbl> 16.46, 17.02, 18.61, 19.44, 17.02, 20.22, 15.84, 20.00, 22.90, 18…
#> $ vs <dbl> 0, 0, 1, 1, 0, 1, 0, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 0,…
#> $ am <dbl> 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 0, 0,…
#> $ gear <dbl> 4, 4, 4, 3, 3, 3, 3, 4, 4, 4, 4, 3, 3, 3, 3, 3, 3, 4, 4, 4, 3, 3,…
#> $ carb <dbl> 4, 4, 1, 1, 2, 1, 4, 2, 2, 4, 4, 3, 3, 3, 4, 4, 4, 1, 2, 1, 1, 2,…Example 4: Writing to a Stata File
If you wish to export a data frame to a Stata format, you can use haven’s write function:
automobile |> write_dta("automobile-data.dta")rio Package
The rio package complements other data-importing packages by providing a one-stop solution for data import and export in R. It handles a wide variety of file types—CSV, Excel, SPSS, Stata, and more—so you don’t need to remember different functions for each format.
import(): Automatically detects the file type and imports your data into R, simplifying the process of reading data from diverse sources.export(): Export your data frame to various file formats with ease, whether you are creating a CSV file, an Excel workbook, or a file for statistical software.
For further details on the extensive capabilities of the rio package, please refer to the rio documentation.
Example 1: Data Import with rio Package
In this example, we load the telco-customer-churn.csv file from the r-data folder using the import() function. This function automatically detects the file type and loads the data accordingly.
After importing the data, you may wish to perform some basic cleaning. For instance, you might standardise variable names using the clean_names() function from the janitor package:
library(janitor)
# Clean variable names
telco_customer_churn <- telco_customer_churn |>
clean_names()
# Glimpse the structure of the data
telco_customer_churn |>
glimpse()#> Rows: 7,043
#> Columns: 21
#> $ customer_id <chr> "7590-VHVEG", "5575-GNVDE", "3668-QPYBK", "7795-CFOC…
#> $ gender <chr> "Female", "Male", "Male", "Male", "Female", "Female"…
#> $ senior_citizen <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
#> $ partner <chr> "Yes", "No", "No", "No", "No", "No", "No", "No", "Ye…
#> $ dependents <chr> "No", "No", "No", "No", "No", "No", "Yes", "No", "No…
#> $ tenure <int> 1, 34, 2, 45, 2, 8, 22, 10, 28, 62, 13, 16, 58, 49, …
#> $ phone_service <chr> "No", "Yes", "Yes", "No", "Yes", "Yes", "Yes", "No",…
#> $ multiple_lines <chr> "No phone service", "No", "No", "No phone service", …
#> $ internet_service <chr> "DSL", "DSL", "DSL", "DSL", "Fiber optic", "Fiber op…
#> $ online_security <chr> "No", "Yes", "Yes", "Yes", "No", "No", "No", "Yes", …
#> $ online_backup <chr> "Yes", "No", "Yes", "No", "No", "No", "Yes", "No", "…
#> $ device_protection <chr> "No", "Yes", "No", "Yes", "No", "Yes", "No", "No", "…
#> $ tech_support <chr> "No", "No", "No", "Yes", "No", "No", "No", "No", "Ye…
#> $ streaming_tv <chr> "No", "No", "No", "No", "No", "Yes", "Yes", "No", "Y…
#> $ streaming_movies <chr> "No", "No", "No", "No", "No", "Yes", "No", "No", "Ye…
#> $ contract <chr> "Month-to-month", "One year", "Month-to-month", "One…
#> $ paperless_billing <chr> "Yes", "No", "Yes", "No", "Yes", "Yes", "Yes", "No",…
#> $ payment_method <chr> "Electronic check", "Mailed check", "Mailed check", …
#> $ monthly_charges <dbl> 29.85, 56.95, 53.85, 42.30, 70.70, 99.65, 89.10, 29.…
#> $ total_charges <dbl> 29.85, 1889.50, 108.15, 1840.75, 151.65, 820.50, 194…
#> $ churn <chr> "No", "No", "Yes", "No", "Yes", "Yes", "No", "No", "…Example 2: Exporting Data with rio Package
After importing your data, it is often necessary to review and adjust variable types. In this example, all character columns (which represent categorical variables) are converted to factors, except for the customer_id column which remains as a character to preserve its unique identifier status. Once the data is transformed (see Chapter 5 for further discussion on data transformation), we filter the data to include only records where dependents is "Yes" and then export the result to an Excel file.
In this manner, rio offers a streamlined process for both importing and exporting data, reducing the need to juggle multiple packages or recall numerous functions.
4.8.4 Web Scraping
Web scraping is the process of extracting data from websites, and R provides several tools to facilitate this task. The ralger package is one such tool, offering a streamlined approach to retrieving and parsing data from web pages. Whether you are collecting data for text analysis, monitoring website updates, or simply gathering information from the web, ralger simplifies the process.
4.8.4.1 Key Features of the ralger Package
Simplified Data Retrieval:
ralgerallows you to quickly download the HTML content of a web page without requiring multiple packages.CSS Selector Support: The package enables you to extract specific elements from a webpage by utilising CSS selectors.
Integrated Parsing Functions: Once the HTML is retrieved,
ralgerprovides functions to parse and manipulate the data efficiently.
Example 1: Extracting a Non-Table HTML Data
When web content is provided in HTML format, you can use the tidy_scrap() function from the ralger package to extract data into a tidy data frame. This function returns a data frame based on the arguments you supply. The key arguments are:
link: The URL of the website you wish to scrape.
nodes: A vector of CSS selectors corresponding to the HTML elements you want to extract. These elements will form the columns of your data frame.
colnames: A vector of names to assign to the columns, which should match the order of the selectors specified in the nodes argument.
clean: If set to
TRUE, the function will clean the tibble’s columns.askRobot: If enabled, the function will consult the site’s
robots.txtfile to verify whether scraping is permitted.
In this example, we will scrape the Hacker News website (https://news.ycombinator.com). We aim to extract a tidy data frame containing the following elements:
The story title.
The site name (if available).
The score of the story.
Additional subline information.
The username of the poster.
Here is how you can achieve this using the tidy_scrap() function with improved column names:
library(ralger)
# Define the URL for Hacker News
url <- "https://news.ycombinator.com/"
# Define the CSS selectors for the elements to extract
nodes <- c(".titleline > a", ".sitestr", ".score", ".subline a+ a", ".hnuser")
# Define descriptive column names for the resulting data frame
colnames <- c("title", "site", "score", "subline", "username")
# Extract the data from all pages using tidy_scrap()
news_data <- tidy_scrap(
link = url,
nodes = nodes,
colnames = colnames,
clean = TRUE
)#> Warning in (function (..., deparse.level = 1) : number of rows of result is not
#> a multiple of vector length (arg 3)news_data#> # A tibble: 30 × 5
#> title site score subline username
#> <chr> <chr> <chr> <chr> <chr>
#> 1 Archival Storage dshr… 142 … 75 com… rbanffy
#> 2 Show HN: OpenTimes – Free travel times between … open… 59 p… 19 com… dfsnow
#> 3 The High Heel Problem simo… 45 p… 5 comm… lehi
#> 4 Deep Learning Is Not So Mysterious or Different arxi… 218 … 56 com… wuubuu
#> 5 Hidden Messages in Emojis and Hacking the US Tr… slam… 62 p… 26 com… nickagl…
#> 6 Alphabet spins out Taara – Internet over lasers x.co… 80 p… 86 com… tadeegan
#> 7 PrintedLabs – 3D printable optical experiment e… uni-… 9 po… discuss aethert…
#> 8 HTTP/3 is everywhere but nowhere http… 281 … 283 co… doener
#> 9 Luthor (YC F24) Is Hiring Ruby on Rails Enginee… ycom… 8 po… discuss ftr1200
#> 10 Show HN: Cascii – A portable ASCII diagram buil… gith… 829 … 325 co… radeeya…
#> # ℹ 20 more rowsIn this code:
The
tidy_scrap()function is called with the Hacker News URL.-
The nodes argument specifies the CSS selectors for the elements we wish to extract. The nodes vector contains the CSS selectors:
“.titleline > a”: extracts the story title,
“.sitestr”: extracts the site name,
“.score”: extracts the story score,
“.subline a+ a”: extracts additional subline information,
“.hnuser”: extracts the username.
The colnames argument assigns clear and descriptive names to the columns—namely
"title","site","score","subline", and"username".The function returns a tidy data frame in which all columns are of character class; you may convert these to other types as required for your analysis.
If you wish to scrape multiple list pages, you can use tidy_scrap() in conjunction with paste0(). Suppose you want to scrape pages 1 through 5 of Hacker News:
# Define the URL for Hacker News
url <- "https://news.ycombinator.com/"
# Create a vector of URLs for pages 1 to 5
links <- paste0(url, "?p=", seq(1, 5, 1))
# Extract the data from all pages using tidy_scrap()
news_data <- tidy_scrap(
link = links,
nodes = nodes,
colnames = colnames
)#> Undefined Error: Error in open.connection(x, "rb"): Could not resolve host: news.ycombinator.com#> Warning in (function (..., deparse.level = 1) : number of rows of result is not
#> a multiple of vector length (arg 2)news_data#> # A tibble: 150 × 5
#> title site score subline username
#> <chr> <chr> <chr> <chr> <chr>
#> 1 Archival Storage dshr… <NA> 75 com… rbanffy
#> 2 Show HN: OpenTimes – Free travel times between … open… <NA> 19 com… dfsnow
#> 3 The High Heel Problem simo… <NA> 5 comm… lehi
#> 4 Deep Learning Is Not So Mysterious or Different arxi… <NA> 56 com… wuubuu
#> 5 Hidden Messages in Emojis and Hacking the US Tr… slam… <NA> 26 com… nickagl…
#> 6 Alphabet spins out Taara – Internet over lasers x.co… <NA> 86 com… tadeegan
#> 7 PrintedLabs – 3D printable optical experiment e… uni-… <NA> discuss aethert…
#> 8 HTTP/3 is everywhere but nowhere http… <NA> 283 co… doener
#> 9 Luthor (YC F24) Is Hiring Ruby on Rails Enginee… ycom… <NA> discuss ftr1200
#> 10 Show HN: Cascii – A portable ASCII diagram buil… gith… <NA> 325 co… radeeya…
#> # ℹ 140 more rowsSince Hacker News is a dynamic website, the content may change over time. Therefore, if you rerun the same lines of code at a later time, the extracted results may differ from those obtained previously.
Example 2: Extracting an HTML Table
The ralger package also includes a function called table_scrap() for extracting HTML tables from a web page. Suppose you want to extract an HTML table from a page listing the highest lifetime gross revenues in the cinema industry. You can use the following code:
# Extract an HTML table from the specified URL
url <- "https://www.boxofficemojo.com/chart/top_lifetime_gross/?area=XWW"
lifetime_gross <- table_scrap(link = url)#> Warning: The `fill` argument of `html_table()` is deprecated as of rvest 1.0.0.
#> ℹ An improved algorithm fills by default so it is no longer needed.
#> ℹ The deprecated feature was likely used in the rvest package.
#> Please report the issue at <https://github.com/tidyverse/rvest/issues>.# Display the extracted table
lifetime_gross#> # A tibble: 200 × 4
#> Rank Title `Lifetime Gross` Year
#> <int> <chr> <chr> <int>
#> 1 1 Avatar $2,923,710,708 2009
#> 2 2 Avengers: Endgame $2,799,439,100 2019
#> 3 3 Avatar: The Way of Water $2,320,250,281 2022
#> 4 4 Titanic $2,264,812,968 1997
#> 5 5 Ne Zha 2 $2,084,181,921 2025
#> 6 6 Star Wars: Episode VII - The Force Awakens $2,071,310,218 2015
#> 7 7 Avengers: Infinity War $2,052,415,039 2018
#> 8 8 Spider-Man: No Way Home $1,952,732,181 2021
#> 9 9 Inside Out 2 $1,698,863,816 2024
#> 10 10 Jurassic World $1,671,537,444 2015
#> # ℹ 190 more rowsIf you are dealing with a web page that contains multiple HTML tables, you can use the choose argument with table_scrap() to target a specific table. For more advanced use cases and customisation options, please refer to the package documentation.
By incorporating web scraping into your data analysis workflow, you can dynamically collect data from the web and integrate it with your existing analyses, thereby broadening the scope of your data-driven insights.
4.8.5 Bringing It All Together
When working on a project, it is best practice to organise your data and code within a dedicated RStudio project. Store your data files (be they flat files, spreadsheets, or labelled files) in a folder (for example, data/), and use relative paths when reading and writing data. This approach promotes reproducibility and clarity. Let us now practise importing data using the gapminder.csv file.
Before We Begin:
Create a Directory
Create a new folder on your desktop calledExperiment 4.3.Download Data
Visit Google Drive to download ther-datafolder (refer to Appendix B.1 for additional information). Once downloaded, unzip the folder and move it into yourExperiment 4.3folder.-
Create an RStudio Project
Open RStudio and set up a new project as follows:- Go to File > New Project.
- Select Existing Directory and navigate to your
Experiment 4.3folder.
This project setup will organise your work and ensure that everything related to this experiment is contained in one place.
- Go to File > New Project.
Your project structure should resemble the one shown in Figure 4.12:
Now, let us import the gapminder.csv file from the r-data folder into R using the tidyverse package for convenient data manipulation and visualisation:
#> Rows: 1704 Columns: 6
#> ── Column specification ────────────────────────────────────────────────────────
#> Delimiter: ","
#> chr (2): country, continent
#> dbl (4): year, lifeExp, pop, gdpPercap
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.# Explore the data
names(gapminder)#> [1] "country" "continent" "year" "lifeExp" "pop" "gdpPercap"dim(gapminder)#> [1] 1704 6head(gapminder)#> # A tibble: 6 × 6
#> country continent year lifeExp pop gdpPercap
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Afghanistan Asia 1952 28.8 8425333 779.
#> 2 Afghanistan Asia 1957 30.3 9240934 821.
#> 3 Afghanistan Asia 1962 32.0 10267083 853.
#> 4 Afghanistan Asia 1967 34.0 11537966 836.
#> 5 Afghanistan Asia 1972 36.1 13079460 740.
#> 6 Afghanistan Asia 1977 38.4 14880372 786.summary(gapminder)#> country continent year lifeExp
#> Length:1704 Length:1704 Min. :1952 Min. :23.60
#> Class :character Class :character 1st Qu.:1966 1st Qu.:48.20
#> Mode :character Mode :character Median :1980 Median :60.71
#> Mean :1980 Mean :59.47
#> 3rd Qu.:1993 3rd Qu.:70.85
#> Max. :2007 Max. :82.60
#> pop gdpPercap
#> Min. :6.001e+04 Min. : 241.2
#> 1st Qu.:2.794e+06 1st Qu.: 1202.1
#> Median :7.024e+06 Median : 3531.8
#> Mean :2.960e+07 Mean : 7215.3
#> 3rd Qu.:1.959e+07 3rd Qu.: 9325.5
#> Max. :1.319e+09 Max. :113523.1After running this script, your dataset should be loaded into RStudio and ready for exploration. Your script should resemble the one in Figure 4.13:
This setup ensures that everything required for your analysis is neatly organised, reproducible, and ready for you to begin analysing the gapminder data in R.
4.8.6 Practice Quiz 4.3
Question 1:
Which package is commonly used to read CSV files into R as tibbles?
-
readxl
-
haven
-
readr
writexl
Question 2:
If you need to import an Excel file, which function would you likely use?
Question 3:
Which package would you use to easily handle a wide variety of data formats without memorising specific functions for each?
-
rio
-
haven
-
janitor
readxl
Question 4:
After cleaning and analysing your data, which function would you use to write the results to a CSV file?
4.8.7 Exercise 4.3.1: Medical Insurance Data
In this exercise, you’ll explore the medical-insurance.xlsx file located in the r-data folder. You can download this file from Google Drive. This dataset contains medical insurance information for various individuals. Below is an overview of each column:
User ID: A unique identifier for each individual.
Gender: The individual’s gender (‘Male’ or ‘Female’).
Age: The age of the individual in years.
AgeGroup: The age bracket the individual falls into.
Estimated Salary: An estimate of the individual’s yearly salary.
Purchased: Indicates whether the individual has purchased medical insurance (1 for Yes, 0 for No).
Your Tasks:
-
Importing and Basic Handling:
- Create a new script and import the data from the Excel file.
- How would you import this data if it’s in SPSS format?
- Use the
clean_names()function from thejanitorpackage to make variable names consistent and easy to work with. - Can you display the first three rows of the dataset?
- How many rows and columns does the dataset have?
-
Understanding the Data:
- What are the column names in the dataset?
- Can you identify the data types of each column?
-
Basic Descriptive Statistics:
- What is the average age of the individuals in the dataset?
- What’s the range of the estimated salaries?
4.9 Reflective Summary
In Lab 4, you have acquired essential skills to enhance your efficiency and effectiveness as an R programmer:
Installing and Loading Packages: You learned how to find, install, and load packages from CRAN and external repositories like GitHub.
Reproducible Workflows with RStudio Projects: You discovered the importance of organizing your work within RStudio Projects.
Importing and Exporting Data: You practiced importing and exporting data in various formats (CSV, Excel, SPSS) using packages like
readr,readxl, andhaven.
These skills are fundamental for efficient data analysis, helping you manage diverse data sources, maintain integrity in your analyses, and collaborate more effectively. Congratulations on progressing as an R programmer and data analyst!
In the next lab, we’ll delve into data transformation where will reshape raw data into a more useful format for data analysis.
It is common for a package to print out messages when you load it. These messages often include information about the package version, attached packages, or important notes from the authors. For example, when you load the tidyverse package. If you prefer to suppress these messages, you can use the
suppressMessages()function:suppressMessages(library(tidyverse))↩︎If you or your collaborators rely on spreadsheets to organise data, we highly recommend the paper “Data Organization in Spreadsheets” by Karl Broman and Kara Woo: https://doi.org/10.1080/00031305.2017.1375989. It provides valuable guidance on best practices and efficient data structuring techniques.↩︎
A tibble is a modern version of an R data frame that provides cleaner printing and more consistent behavior, especially within the tidyverse ecosystem.↩︎