bar(foo(data))5 Data Transformation
5.1 Introduction
Welcome to Lab 5! In this lab, we will focus on one of the most important steps in the data analysis process: data transformation. Real-world data rarely arrives in perfect, analysis-ready form. Before generating insights, visualising patterns, or constructing models, we must transform the data: cleaning, reshaping, and summarising it into a more meaningful structure.
In this lab, we will explore:
The native pipe operator
|>to create a smooth, readable pipeline of data operations.The core
dplyrverbs—select(),filter(),mutate(),arrange(), andsummarise()—to efficiently manipulate and refine your data.Strategies for grouping and summarising data to extract patterns and trends.
Techniques for identifying and handling missing values responsibly.
These transformations form a vital step in any data workflow, ensuring that your datasets are well-prepared for subsequent analysis or visualisation.
5.2 Learning Objectives
By the end of this lab, you will be able to:
Streamline Your Code Using the Pipe Operator
|>
Connect multiple data operations into a logical sequence, enhancing code readability and reducing nesting complexity.Perform Key Data Transformation Tasks Using
dplyr
Utilise functions likeselect(),filter(),arrange(),mutate(), andsummarise()toreshape, refine, and improve datasets for analysis.Gain Insights Through Summarisation and Grouping
Group data and apply aggregation functions to extract meaningful summaries and trends.Handle and Impute Missing Data
Identify missing values in your datasets, understand their impact, and apply suitable strategies (e.g., removal, mean/median imputation) to maintain data integrity.Prepare Data for Analysis and Visualisation
Clean and structure your datasets so they are ready for modelling, plotting, and communicating results effectively.
By completing this lab, you will master data transformation and become more confident in dealing with messy, real-world datasets and advancing towards deeper analyses.
5.3 Prerequisites
Before starting this lab, you should have:
5.4 What is Data Transformation?
Data transformation is the process of reshaping raw data into a more useful format. Real-world data is rarely perfect for immediate analysis. It often contains extra variables, missing values, or is structured in a way that is not conducive to answering your research questions. Data transformation includes:
Selecting relevant parts of the dataset: Focusing on the rows and columns you actually need.
Creating new variables: Deriving meaningful metrics from existing data.
Summarising information: Computing averages, totals, or other aggregate statistics to distil complex information into digestible summaries.
Organising data into a meaningful structure: Arranging your dataset so that the relationships between variables and observations are clear.
You can think of data transformation like preparing ingredients before cooking: you wash, chop, and measure everything out so that when you start cooking, you can focus on creating your dish without interruptions or confusion.
5.5 Real-World Scenario: Preparing Data for Analysis
Imagine that you work as a data analyst at a wildlife research centre. You have received a raw dataset containing information on various species: their body measurements, diets, sleep patterns, and more. Before you can analyse ecological relationships, test hypotheses, or build models, you need to clean and organise this data. Using the techniques in this lab, you can:
Select only the columns necessary for your investigation.
Filter out rows that are not relevant to your study.
Mutate the dataset to create new metrics or correct errors.
Arrange the rows to highlight the largest or smallest values.
Summarise the data by group to identify patterns by species or habitat.
Handle missing values so that they do not distort your findings.
By applying these transformations, you ensure that your data is analysis-ready.
5.6 Experiment 5.1: The Pipe Operator |>
One of the best tools to simplify your R code is the pipe operator. Traditionally, the <%> operator from the magrittr package has been widely used for this purpose. However, starting from R version 4.1.0, R introduced a native pipe operator |>. The pipe operator allows you to chain functions together in a linear, logical sequence, rather than nesting them inside one another. Using pipes makes your code more readable and helps you think through your data transformations step-by-step.
In this lab, we will be using the base pipe operator |>, which functions similarly to the magrittr <%> operator1. Imagine you have data frame, data, and you want to perform multiple operations on it, such as applying functions foo and bar in sequence. Without a pipe, you might write as:
This is harder to read than:
data |>
foo() |>
bar()In the piped version, you start with data, then say “and then apply foo(),” and then “and then apply bar(),” which feels more intuitive and mirrors how we naturally describe processes in words.
To configure RStudio to insert the base pipe operator |> instead of %>% when pressing Ctrl/Cmd + Shift + M, navigate to the Tools menu, select Global Options…, then go to the Code section. In the Code options, check the box labelled Use native pipe operator, |> (requires R 4.1+).
How Does the Pipe Operator Work?
The pipe operator |> takes the output of one function and passes it as the first argument to the next function.
Example 1
For instance, consider:
iris |> head()#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 5.1 3.5 1.4 0.2 setosa
#> 2 4.9 3.0 1.4 0.2 setosa
#> 3 4.7 3.2 1.3 0.2 setosa
#> 4 4.6 3.1 1.5 0.2 setosa
#> 5 5.0 3.6 1.4 0.2 setosa
#> 6 5.4 3.9 1.7 0.4 setosaThis is exactly the same as:
head(iris)#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 5.1 3.5 1.4 0.2 setosa
#> 2 4.9 3.0 1.4 0.2 setosa
#> 3 4.7 3.2 1.3 0.2 setosa
#> 4 4.6 3.1 1.5 0.2 setosa
#> 5 5.0 3.6 1.4 0.2 setosa
#> 6 5.4 3.9 1.7 0.4 setosaExample 2:
Here’s another example combining multiple functions:
This sequence is equivalent to the nested version:
How does using the pipe operator enhance clarity, compared to nested function calls, especially when performing multiple operations on the same dataset?
By using pipes, you avoid writing nested code and make the flow of your data transformation much clearer.
5.6.1 Practice Quiz 5.1
Question 1:
What is the primary purpose of the pipe operator (|> or %>%) in R?
- To run code in parallel.
- To nest functions inside one another.
- To pass the output of one function as the input to the next, improving code readability.
- To automatically clean missing data.
Question 2:
Consider the following R code snippets:
numbers <- c(2, 4, 6)
# Nested function version:
result1 <- round(sqrt(sum(numbers)))
# Pipe operator version:
result2 <- numbers |> sum() |> sqrt() |> round()For a new R learner, is the pipe operator version generally more readable than the nested function version?
- True
- False
Question 3:
What is the output of the following R code?
- 10
- 15
- 5
- 30
Question 4:
Which of the following code snippets correctly uses the pipe operator to apply the sqrt() function to the sum of numbers from 1 to 4?
-
sqrt(sum(1:4))
-
1:4 |> sum() |> sqrt()
-
sum(1:4) |> sqrt
1:4 |> sqrt() |> sum()
Question 5:
What will be the output of the following code?
result <- letters
result |> head(3)-
c("a", "b", "c")
-
c("x", "y", "z")
-
c("A", "B", "C")
- An error is thrown.
5.7 Experiment 5.2: Data Manipulation with dplyr
In data analysis, we often encounter datasets that aren’t in the ideal format for our needs. Data comes in all shapes and sizes, and making sense of it requires effective manipulation. This is where data manipulation becomes essential—a fundamental skill that allows you to transform and summarize data efficiently.
The dplyr package, part of the tidyverse, is designed to make data manipulation in R more approachable, efficient, and intuitive. Think of dplyr as your Swiss Army knife for taming messy datasets. It simplifies tasks like filtering, summarizing, grouping, and transforming data. The best part? Its syntax is easy to read and write, almost like having a conversation with your data.
Why Use dplyr?
Simplicity: Provides straightforward functions that are easy to learn and remember, lowering the barrier to effective data manipulation.
Efficiency: Optimized for performance, it handles large datasets swiftly, saving you time and computational resources.
Readability: Code written with dplyr is often more readable and easier to maintain, which is especially beneficial when collaborating with others or revisiting your own work.
Integration: Works seamlessly with other tidyverse packages like ggplot2 and tidyr, allowing for a cohesive and efficient data analysis workflow.
Getting Started
First, ensure you have the dplyr package installed and loaded. If you haven’t installed it yet, you can install the tidyverse, which includes dplyr.
# Install the tidyverse package (if not already installed)
install.packages("tidyverse")
# Load the tidyverse package
library(tidyverse)Core dplyr Verbs
The core functions in dplyr are often referred to as “verbs” because they describe actions you perform on your data:
select(): Choose variables (columns) based on their names or column positions.mutate(): Create new columns or modify existing ones.filter(): Select rows based on specific conditions.arrange(): Reorder rows based on column values.summarise(): Reduce multiple values down to a summary statistic.group_by(): Group data by one or more variables for grouped operations.
When summarise() is paired with group_by(), it allows you to get a summary row for each group in the data frame.
Additional useful functions include:
rename(): Rename columns.distinct(): Find unique rows.count(): Count unique values of a variable.
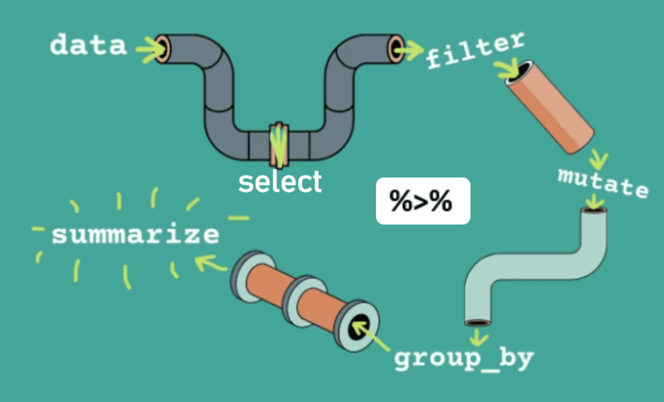
Using Pipes with dplyr functions
One of the key strengths of dplyr is its ability to integrate seamlessly with the pipe operator (%>% or |>), enabling a clean, readable, and intuitive workflow for data manipulation. As illustrated in Figure 5.4, the pipe operator acts as a connector, allowing you to chain multiple dplyr functions in a logical sequence. This approach makes your code easier to read and follow, mirroring the flow of a conversation about your data.
5.7.1 Working with the dplyr Verbs
Let’s take a deeper dive into each dplyr verb and understand not just how to use them but also what makes them powerful. Remember, the five core verbs—filter(), select(), mutate(), arrange(), and summarize()—are like tools in a toolbox. Each has a specific purpose, but together, they allow you to transform data seamlessly.
Example Datasets
We’ll start our exploration by working with two fascinating datasets: the penguins dataset from the palmerpenguins package2 and the msleep dataset from the ggplot2 package. These datasets provide rich, real-world data that will help you practice and apply data manipulation in this book.
- The
penguinsDataset
The penguins dataset3 contains detailed body measurements for 344 penguins from three different species—Adélie, Chinstrap, and Gentoo—found on three islands in the Palmer Archipelago of Antarctica. This dataset includes variables such as:
Species: The penguin species.
Island: The island where each penguin was observed.
Bill Length and Depth: Measurements of the penguin’s bill (beak).
Flipper Length: The length of the penguin’s flippers.
Body Mass: The weight of the penguin.
Sex: The gender of the penguin.
#> Rows: 344
#> Columns: 8
#> $ species <fct> Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Adel…
#> $ island <fct> Torgersen, Torgersen, Torgersen, Torgersen, Torgerse…
#> $ bill_length_mm <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.3, 38.9, 39.2, 34.1, …
#> $ bill_depth_mm <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.6, 17.8, 19.6, 18.1, …
#> $ flipper_length_mm <int> 181, 186, 195, NA, 193, 190, 181, 195, 193, 190, 186…
#> $ body_mass_g <int> 3750, 3800, 3250, NA, 3450, 3650, 3625, 4675, 3475, …
#> $ sex <fct> male, female, female, NA, female, male, female, male…
#> $ year <int> 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007…- The
msleepDataset
Our second dataset, msleep, comes from the ggplot2 package and contains information on the sleep habits of 83 different mammals. This dataset includes 11 variables, such as:
name: The common name of the mammal.
genus: The taxonomic genus of the mammal.
-
vore: The dietary category of the mammal. Possible values include:
"carni": Carnivore (meat-eating)"herbi": Herbivore (plant-eating)"omni": Omnivore (eating both plants and meat)"insecti": Insectivore (eating insects)
order: The taxonomic order to which the mammal belongs (e.g., Primates, Carnivora).
-
conservation: The conservation status of the species, indicating its level of threat or endangerment. Possible values include:
"lc": Least Concern"nt": Near Threatened"vu": Vulnerable"en": Endangered"cr": Critically Endangered"domesticated": Domesticated species
sleep total: Total amount of sleep per day (in hours).
sleep rem: Amount of REM sleep per day.
sleep cycle: Length of the sleep cycle.
awake: The number of hours the mammal spends awake each day (calculated as
24 - sleep_total).brain weight: The brain weight of the animal.
body weight: The body weight of the animal.
#> Rows: 83
#> Columns: 11
#> $ name <chr> "Cheetah", "Owl monkey", "Mountain beaver", "Greater shor…
#> $ genus <chr> "Acinonyx", "Aotus", "Aplodontia", "Blarina", "Bos", "Bra…
#> $ vore <chr> "carni", "omni", "herbi", "omni", "herbi", "herbi", "carn…
#> $ order <chr> "Carnivora", "Primates", "Rodentia", "Soricomorpha", "Art…
#> $ conservation <chr> "lc", NA, "nt", "lc", "domesticated", NA, "vu", NA, "dome…
#> $ sleep_total <dbl> 12.1, 17.0, 14.4, 14.9, 4.0, 14.4, 8.7, 7.0, 10.1, 3.0, 5…
#> $ sleep_rem <dbl> NA, 1.8, 2.4, 2.3, 0.7, 2.2, 1.4, NA, 2.9, NA, 0.6, 0.8, …
#> $ sleep_cycle <dbl> NA, NA, NA, 0.1333333, 0.6666667, 0.7666667, 0.3833333, N…
#> $ awake <dbl> 11.9, 7.0, 9.6, 9.1, 20.0, 9.6, 15.3, 17.0, 13.9, 21.0, 1…
#> $ brainwt <dbl> NA, 0.01550, NA, 0.00029, 0.42300, NA, NA, NA, 0.07000, 0…
#> $ bodywt <dbl> 50.000, 0.480, 1.350, 0.019, 600.000, 3.850, 20.490, 0.04…
5.7.2 select() – Picking Specific Columns
The select() function allows you to extract specific columns from a dataset, focusing on only the features relevant to your analysis. It’s like creating a spotlight for just the features you need.
Key Points:
-
Select Columns by Name or Position:
You can specify columns by their names or their positions (e.g., 1, 2, 3).
This is useful when you only need a few specific columns from a large dataset.
-
Use Helper Functions for Pattern Matching:
starts_with("prefix"): Selects columns whose names begin with a specific prefix.ends_with("suffix"): Selects columns whose names end with a specific suffix.contains("substring"): Selects columns whose names contain a specific substring.
-
Deselect Columns Using the Minus (
-) Sign:You can drop columns by prefixing their names, indices, or helper functions with
-.This is useful when you want to keep most columns but exclude specific ones.
-
- This allows you to select columns programmatically or apply operations to subsets of columns.
5.7.2.1 Selecting Columns by Name:
Suppose we want to extract the name, vore, and sleep_total from the msleep data:
msleep |>
select(name, sleep_total, vore)#> # A tibble: 83 × 3
#> name sleep_total vore
#> <chr> <dbl> <chr>
#> 1 Cheetah 12.1 carni
#> 2 Owl monkey 17 omni
#> 3 Mountain beaver 14.4 herbi
#> 4 Greater short-tailed shrew 14.9 omni
#> 5 Cow 4 herbi
#> 6 Three-toed sloth 14.4 herbi
#> 7 Northern fur seal 8.7 carni
#> 8 Vesper mouse 7 <NA>
#> 9 Dog 10.1 carni
#> 10 Roe deer 3 herbi
#> # ℹ 73 more rows5.7.2.2 Selecting Columns by Position:
You can use the position of columns to select them, for example, the first three columns:
msleep |>
select(1:3)#> # A tibble: 83 × 3
#> name genus vore
#> <chr> <chr> <chr>
#> 1 Cheetah Acinonyx carni
#> 2 Owl monkey Aotus omni
#> 3 Mountain beaver Aplodontia herbi
#> 4 Greater short-tailed shrew Blarina omni
#> 5 Cow Bos herbi
#> 6 Three-toed sloth Bradypus herbi
#> 7 Northern fur seal Callorhinus carni
#> 8 Vesper mouse Calomys <NA>
#> 9 Dog Canis carni
#> 10 Roe deer Capreolus herbi
#> # ℹ 73 more rows5.7.2.3 Selecting Columns Using Patterns:
- Columns Starting with a Prefix: Select columns starting with “sleep”:
msleep |>
select(starts_with("sleep"))#> # A tibble: 83 × 3
#> sleep_total sleep_rem sleep_cycle
#> <dbl> <dbl> <dbl>
#> 1 12.1 NA NA
#> 2 17 1.8 NA
#> 3 14.4 2.4 NA
#> 4 14.9 2.3 0.133
#> 5 4 0.7 0.667
#> 6 14.4 2.2 0.767
#> 7 8.7 1.4 0.383
#> 8 7 NA NA
#> 9 10.1 2.9 0.333
#> 10 3 NA NA
#> # ℹ 73 more rows- Columns Ending with a Suffix: Select columns ending with “wt”:
#> # A tibble: 83 × 2
#> brainwt bodywt
#> <dbl> <dbl>
#> 1 NA 50
#> 2 0.0155 0.48
#> 3 NA 1.35
#> 4 0.00029 0.019
#> 5 0.423 600
#> 6 NA 3.85
#> 7 NA 20.5
#> 8 NA 0.045
#> 9 0.07 14
#> 10 0.0982 14.8
#> # ℹ 73 more rows- Columns Containing a Substring: Select columns that contain the word “con”:
5.7.2.4 Deselect Columns Using the Minus Sign:
If you want to keep all columns except name and sleep_total:
#> # A tibble: 83 × 9
#> genus vore order conservation sleep_rem sleep_cycle awake brainwt bodywt
#> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Acinon… carni Carn… lc NA NA 11.9 NA 50
#> 2 Aotus omni Prim… <NA> 1.8 NA 7 0.0155 0.48
#> 3 Aplodo… herbi Rode… nt 2.4 NA 9.6 NA 1.35
#> 4 Blarina omni Sori… lc 2.3 0.133 9.1 0.00029 0.019
#> 5 Bos herbi Arti… domesticated 0.7 0.667 20 0.423 600
#> 6 Bradyp… herbi Pilo… <NA> 2.2 0.767 9.6 NA 3.85
#> 7 Callor… carni Carn… vu 1.4 0.383 15.3 NA 20.5
#> 8 Calomys <NA> Rode… <NA> NA NA 17 NA 0.045
#> 9 Canis carni Carn… domesticated 2.9 0.333 13.9 0.07 14
#> 10 Capreo… herbi Arti… lc NA NA 21 0.0982 14.8
#> # ℹ 73 more rowsYou can also use helper functions to deselect columns. For instance, drop all columns starting with “sleep”
msleep |>
select(-starts_with("sleep"))#> # A tibble: 83 × 8
#> name genus vore order conservation awake brainwt bodywt
#> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl>
#> 1 Cheetah Acin… carni Carn… lc 11.9 NA 50
#> 2 Owl monkey Aotus omni Prim… <NA> 7 0.0155 0.48
#> 3 Mountain beaver Aplo… herbi Rode… nt 9.6 NA 1.35
#> 4 Greater short-tailed s… Blar… omni Sori… lc 9.1 0.00029 0.019
#> 5 Cow Bos herbi Arti… domesticated 20 0.423 600
#> 6 Three-toed sloth Brad… herbi Pilo… <NA> 9.6 NA 3.85
#> 7 Northern fur seal Call… carni Carn… vu 15.3 NA 20.5
#> 8 Vesper mouse Calo… <NA> Rode… <NA> 17 NA 0.045
#> 9 Dog Canis carni Carn… domesticated 13.9 0.07 14
#> 10 Roe deer Capr… herbi Arti… lc 21 0.0982 14.8
#> # ℹ 73 more rows
5.7.2.5 Using where() with select()
The where() helper function enables you to dynamically select or modify columns in your data. When used with select(), it allows you to programmatically choose columns based on specific criteria. For example, to select all numeric columns, you can use where(is.numeric):
#> # A tibble: 83 × 6
#> sleep_total sleep_rem sleep_cycle awake brainwt bodywt
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 12.1 NA NA 11.9 NA 50
#> 2 17 1.8 NA 7 0.0155 0.48
#> 3 14.4 2.4 NA 9.6 NA 1.35
#> 4 14.9 2.3 0.133 9.1 0.00029 0.019
#> 5 4 0.7 0.667 20 0.423 600
#> 6 14.4 2.2 0.767 9.6 NA 3.85
#> 7 8.7 1.4 0.383 15.3 NA 20.5
#> 8 7 NA NA 17 NA 0.045
#> 9 10.1 2.9 0.333 13.9 0.07 14
#> 10 3 NA NA 21 0.0982 14.8
#> # ℹ 73 more rows
5.7.3 mutate() – Creating or Modifying Columns
The mutate() function is like a magic wand for adding new variables or transforming existing ones. Use it whenever you need to derive new information from your dataset.
Key Points:
You can add as many new columns as you need.
Existing columns can be modified by overwriting them.
5.7.3.1 Creating a New Column:
Sleep is a vital physiological process, but its duration varies widely among mammals4. Understanding how sleep duration relates to body weight could reveal insights into the metabolic and ecological factors influencing sleep. For example:
Larger mammals may have lower sleep-to-body-weight ratios due to their lower metabolic rates relative to body size.
Smaller mammals might have higher sleep-to-body-weight ratios, potentially linked to their higher metabolic demands.
To explore this, we calculate the ratio of total sleep (sleep_total) to body weight (bodywt) for each species in the msleep dataset:
#> # A tibble: 83 × 5
#> name vore sleep_total bodywt sleep_to_weight
#> <chr> <chr> <dbl> <dbl> <dbl>
#> 1 Cheetah carni 12.1 50 0.242
#> 2 Owl monkey omni 17 0.48 35.4
#> 3 Mountain beaver herbi 14.4 1.35 10.7
#> 4 Greater short-tailed shrew omni 14.9 0.019 784.
#> 5 Cow herbi 4 600 0.00667
#> 6 Three-toed sloth herbi 14.4 3.85 3.74
#> 7 Northern fur seal carni 8.7 20.5 0.425
#> 8 Vesper mouse <NA> 7 0.045 156.
#> 9 Dog carni 10.1 14 0.721
#> 10 Roe deer herbi 3 14.8 0.203
#> # ℹ 73 more rowsThis ratio provides a standardised measure to compare sleep duration across species with varying body sizes.
In the same msleep dataset, we suspect that any brain weight greater than 4 is likely an error. To address this, we:
Exclude these suspected outliers by replacing them with
NA.Retain valid brain weight values for analysis.
msleep |>
select(name, brainwt) |>
# Replace brainwt > 4 with NA
mutate(brainwt_corrected = ifelse(brainwt > 4, NA, brainwt)) |>
# Sort by original brain weight in descending order
arrange(desc(brainwt))#> # A tibble: 83 × 3
#> name brainwt brainwt_corrected
#> <chr> <dbl> <dbl>
#> 1 African elephant 5.71 NA
#> 2 Asian elephant 4.60 NA
#> 3 Human 1.32 1.32
#> 4 Horse 0.655 0.655
#> 5 Chimpanzee 0.44 0.44
#> 6 Cow 0.423 0.423
#> 7 Donkey 0.419 0.419
#> 8 Gray seal 0.325 0.325
#> 9 Baboon 0.18 0.18
#> 10 Pig 0.18 0.18
#> # ℹ 73 more rowsselect(name, brainwt): Focuses on the columns relevant to this analysis: the species’ name and their brain weight.-
mutate(brainwt_corrected = ifelse(brainwt > 4, NA, brainwt)):Uses
ifelse()to create a new column,brainwt_corrected, where anybrainwtabove 4 is replaced withNA.Values 4 or below remain unchanged.
-
arrange(desc(brainwt)):- Sorts the data by the original
brainwtin descending order, making it easier to verify the replaced outliers. (You will soon learn more about the arrange() function.)
- Sorts the data by the original
To better understand patterns in sleep behaviour, it can be helpful to categorize species into discrete groups based on their sleep duration. This makes it easier to group species for further analysis, or explore ecological or evolutionary hypotheses.
For example, we can categorize species as:
“Long sleepers”: Those that sleep more than 9 hours per day.
“Short sleepers”: Those that sleep 9 hours or less per day.
We achieve this categorization using the ifelse() function, which allows us to transform the continuous numeric variable sleep_total into a categorical variable, sleep_category.
msleep |>
select(name, vore, sleep_total) |>
mutate(sleep_category = ifelse(sleep_total > 9, "long", "short"))#> # A tibble: 83 × 4
#> name vore sleep_total sleep_category
#> <chr> <chr> <dbl> <chr>
#> 1 Cheetah carni 12.1 long
#> 2 Owl monkey omni 17 long
#> 3 Mountain beaver herbi 14.4 long
#> 4 Greater short-tailed shrew omni 14.9 long
#> 5 Cow herbi 4 short
#> 6 Three-toed sloth herbi 14.4 long
#> 7 Northern fur seal carni 8.7 short
#> 8 Vesper mouse <NA> 7 short
#> 9 Dog carni 10.1 long
#> 10 Roe deer herbi 3 short
#> # ℹ 73 more rowsThe ifelse() function is a handy tool for converting a numeric column into a categorical (or discrete) one. As explained earlier, ifelse() works by taking three arguments: a logical condition, a value to return if the condition is TRUE, and a value to return if the condition is FALSE. This makes it an efficient way to create new variables or modify existing ones based on specific criteria.
This categorization opens the door to exploring broader scientific questions:
-
What ecological or metabolic factors correlate with sleep duration?
- For example, are “long sleepers” more likely to be predators, herbivores, or omnivores?
-
Do body size or brain size influence sleep duration?
- Smaller animals may tend to sleep more to conserve energy, while larger animals might sleep less due to lower relative metabolic rates.
-
Are long sleepers more prevalent in certain habitats?
- Does living in safer environments allow for extended sleep?
5.7.3.2 Using mutate() and case_when():
Body weight is often an important ecological indicator, and mammals can be classified into the following categories based on their weight:
Heavy: Body weight > 50 kg
Medium: Body weight > 10 kg but ≤ 50 kg
Light: Body weight ≤ 10 kg
When assigning these categories, the case_when() function provides a more elegant and readable solution compared to using nested ifelse() statements. It allows you to handle multiple conditions cleanly and intuitively. Here’s how it can be used:
msleep |>
select(name, sleep_total, bodywt) |>
mutate(
bodywt_category = case_when(
bodywt > 50 ~ "heavy",
bodywt > 10 ~ "medium",
TRUE ~ "light" # Default for remaining cases
)
)#> # A tibble: 83 × 4
#> name sleep_total bodywt bodywt_category
#> <chr> <dbl> <dbl> <chr>
#> 1 Cheetah 12.1 50 medium
#> 2 Owl monkey 17 0.48 light
#> 3 Mountain beaver 14.4 1.35 light
#> 4 Greater short-tailed shrew 14.9 0.019 light
#> 5 Cow 4 600 heavy
#> 6 Three-toed sloth 14.4 3.85 light
#> 7 Northern fur seal 8.7 20.5 medium
#> 8 Vesper mouse 7 0.045 light
#> 9 Dog 10.1 14 medium
#> 10 Roe deer 3 14.8 medium
#> # ℹ 73 more rowsWe can combine both categorizations into a single dataset to examine potential relationships between sleep behaviour and body weight. For example, we could ask:
- Are “light” mammals more likely to be “short sleepers”?
To create ordered factors for better control in plots or analyses, we use factor() or the forcats package:
msleep |>
select(name, sleep_total, bodywt) |>
mutate(
sleep_category = ifelse(sleep_total > 9, "long", "short"),
bodywt_discr = case_when(
bodywt > 50 ~ "heavy",
bodywt > 10 ~ "medium",
TRUE ~ "light"
),
# Convert to ordered factors
sleep_category = factor(sleep_category, levels = c("short", "long")),
bodywt_discr = factor(bodywt_discr, levels = c("light", "medium", "heavy"))
)#> # A tibble: 83 × 5
#> name sleep_total bodywt sleep_category bodywt_discr
#> <chr> <dbl> <dbl> <fct> <fct>
#> 1 Cheetah 12.1 50 long medium
#> 2 Owl monkey 17 0.48 long light
#> 3 Mountain beaver 14.4 1.35 long light
#> 4 Greater short-tailed shrew 14.9 0.019 long light
#> 5 Cow 4 600 short heavy
#> 6 Three-toed sloth 14.4 3.85 long light
#> 7 Northern fur seal 8.7 20.5 short medium
#> 8 Vesper mouse 7 0.045 short light
#> 9 Dog 10.1 14 long medium
#> 10 Roe deer 3 14.8 short medium
#> # ℹ 73 more rows
5.7.3.3 Calculating Row-Wise Averages Using mutate()
When analysing mammalian sleep data, researchers may want to compare different sleep metrics for each species. For instance:
REM sleep (
sleep_rem) and sleep cycle duration (sleep_cycle) could be combined to calculate a single representative metric, such as the average of the two values.This average can provide a holistic view of each species’ sleep patterns.
However, standard aggregation functions like mean() or sum() operate on entire columns, summarising all observations at once rather than computing values row by row.
To calculate the average of sleep_rem and sleep_cycle for each species, you can use one of the following approaches:
-
Explicit Arithmetic:
(sleep_rem + sleep_cycle) / 2
#> # A tibble: 83 × 4
#> name sleep_rem sleep_cycle avg_sleep
#> <chr> <dbl> <dbl> <dbl>
#> 1 Cheetah NA NA NA
#> 2 Owl monkey 1.8 NA NA
#> 3 Mountain beaver 2.4 NA NA
#> 4 Greater short-tailed shrew 2.3 0.133 1.22
#> 5 Cow 0.7 0.667 0.683
#> 6 Three-toed sloth 2.2 0.767 1.48
#> 7 Northern fur seal 1.4 0.383 0.892
#> 8 Vesper mouse NA NA NA
#> 9 Dog 2.9 0.333 1.62
#> 10 Roe deer NA NA NA
#> # ℹ 73 more rowsmsleep |>
select(name, sleep_rem, sleep_cycle) |>
rowwise() |> # Enable row-wise operations
mutate(avg_sleep = mean(c(sleep_rem, sleep_cycle), na.rm = TRUE)) |>
ungroup()#> # A tibble: 83 × 4
#> name sleep_rem sleep_cycle avg_sleep
#> <chr> <dbl> <dbl> <dbl>
#> 1 Cheetah NA NA NaN
#> 2 Owl monkey 1.8 NA 1.8
#> 3 Mountain beaver 2.4 NA 2.4
#> 4 Greater short-tailed shrew 2.3 0.133 1.22
#> 5 Cow 0.7 0.667 0.683
#> 6 Three-toed sloth 2.2 0.767 1.48
#> 7 Northern fur seal 1.4 0.383 0.892
#> 8 Vesper mouse NA NA NaN
#> 9 Dog 2.9 0.333 1.62
#> 10 Roe deer NA NA NaN
#> # ℹ 73 more rowsWhen you use rowwise(), it is important to call ungroup() afterwards to remove row-wise grouping and revert to the default, ungrouped state of the data.
5.7.3.4 Using mutate() and across() to Modify Specific Columns
When working with datasets, it is common to encounter situations where you need to apply the same transformation to multiple columns. For instance, you might want to convert units, round numeric values, or standardise text. In such cases, the combination of mutate() and across() from the dplyr package is a powerful tool. These functions allow you to efficiently select and modify subsets of columns, reducing the need for repetitive code.
The across() function is particularly versatile. It enables you to:
Select columns using tidy selection helpers like
starts_with(),contains(), orwhere().Apply a function to each of the selected columns, making it easy to perform bulk operations.
We will explore several practical examples using the msleep dataset, which contains information about the sleep patterns of mammals. These examples will demonstrate how to use mutate() and across() to perform common data transformation tasks.
Example 1: Converting Units for Sleep-Related Columns
The msleep dataset includes several columns related to sleep duration, measured in hours. Suppose you need these values in minutes instead. Rather than manually transforming each column, you can use across() to apply the conversion to all relevant columns at once 5.
msleep |>
select(name, contains("sleep")) |> # Focus on sleep-related columns
mutate(across(contains("sleep"), ~ .x * 60)) # Multiply each value by 60#> # A tibble: 83 × 4
#> name sleep_total sleep_rem sleep_cycle
#> <chr> <dbl> <dbl> <dbl>
#> 1 Cheetah 726 NA NA
#> 2 Owl monkey 1020 108 NA
#> 3 Mountain beaver 864 144 NA
#> 4 Greater short-tailed shrew 894 138 8.00
#> 5 Cow 240 42 40.0
#> 6 Three-toed sloth 864 132 46.0
#> 7 Northern fur seal 522 84 23.0
#> 8 Vesper mouse 420 NA NA
#> 9 Dog 606 174 20.0
#> 10 Roe deer 180 NA NA
#> # ℹ 73 more rows-
select(name, contains("sleep")):- Keeps the
namecolumn (for species identification) and all columns whose names contain the word “sleep”.
- Keeps the
-
mutate(across(contains("sleep"), ~ .x * 60)):across(contains("sleep")): Selects all columns containing “sleep” in their names.The formula ~ .x * 60 converts hours to minutes by multiplying each value by 60.
Uses ~ to define an anonymous function and .x to refer to column values.
This approach is not only concise but also scalable. If new sleep-related columns are added to the dataset, the same code will automatically include them in the transformation.
Example 2: Rounding Numeric Columns
Another common task is rounding numeric values to a specified number of decimal places. For example, you might want to round all numeric columns in the msleep dataset to the nearest integer.
#> # A tibble: 83 × 11
#> name genus vore order conservation sleep_total sleep_rem sleep_cycle awake
#> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Cheet… Acin… carni Carn… lc 12 NA NA 12
#> 2 Owl m… Aotus omni Prim… <NA> 17 2 NA 7
#> 3 Mount… Aplo… herbi Rode… nt 14 2 NA 10
#> 4 Great… Blar… omni Sori… lc 15 2 0 9
#> 5 Cow Bos herbi Arti… domesticated 4 1 1 20
#> 6 Three… Brad… herbi Pilo… <NA> 14 2 1 10
#> 7 North… Call… carni Carn… vu 9 1 0 15
#> 8 Vespe… Calo… <NA> Rode… <NA> 7 NA NA 17
#> 9 Dog Canis carni Carn… domesticated 10 3 0 14
#> 10 Roe d… Capr… herbi Arti… lc 3 NA NA 21
#> # ℹ 73 more rows
#> # ℹ 2 more variables: brainwt <dbl>, bodywt <dbl>mutate(across(where(is.numeric), round)):
where(is.numeric)selects all numeric columns.The
roundfunction is applied to each numeric column, rounding the values to the nearest integer.
This is particularly useful when preparing data for reporting or visualisation, where overly precise values can clutter the output.
Example 3: Scaling Numeric Columns to a Range of 0 to 1
Normalising data is a common preprocessing step in data analysis, particularly when features have different scales. For instance, scaling numeric columns to a range of 0 to 1 ensures comparability across variables. This is achieved using min-max scaling, defined by the formula:
\[ x_{\text{scaled}} = \frac{x - \min(x)}{\max(x) - \min(x)} \]
Where:
\(\min(x)\) is the minimum value in the column.
\(\max(x)\) is the maximum value in the column.
\(x_{\text{scaled}}\) is the normalised value, constrained to the interval \([0, 1]\).
To streamline this process, we encapsulate the formula into a reusable function:
This function:
Ignores missing values (
na.rm = TRUE) to avoidNApropagation.Automatically adapts to each column’s unique range.
Apply the function to all numeric columns in the msleep dataset:
#> # A tibble: 83 × 11
#> name genus vore order conservation sleep_total sleep_rem sleep_cycle awake
#> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Cheet… Acin… carni Carn… lc 0.567 NA NA 0.433
#> 2 Owl m… Aotus omni Prim… <NA> 0.839 0.262 NA 0.161
#> 3 Mount… Aplo… herbi Rode… nt 0.694 0.354 NA 0.306
#> 4 Great… Blar… omni Sori… lc 0.722 0.338 0.0120 0.278
#> 5 Cow Bos herbi Arti… domesticated 0.117 0.0923 0.398 0.883
#> 6 Three… Brad… herbi Pilo… <NA> 0.694 0.323 0.470 0.306
#> 7 North… Call… carni Carn… vu 0.378 0.2 0.193 0.622
#> 8 Vespe… Calo… <NA> Rode… <NA> 0.283 NA NA 0.717
#> 9 Dog Canis carni Carn… domesticated 0.456 0.431 0.157 0.544
#> 10 Roe d… Capr… herbi Arti… lc 0.0611 NA NA 0.939
#> # ℹ 73 more rows
#> # ℹ 2 more variables: brainwt <dbl>, bodywt <dbl>Zero-variance columns: If all values in a column are identical,
max(x) - min(x) = 0, resulting inNaN(division by zero).Missing values: Columns with only
NAvalues will returnNaN.Interpretability: Scaled values retain the relative relationships in the original data.
This transformation ensures all numeric values are proportionally mapped to the same range, simplifying comparisons and improving the performance of machine learning algorithms.
Example 4: Transforming Character Columns
Text data often contains extra whitespace—leading, trailing, or even multiple spaces between words. This can lead to inconsistencies in your analysis. The str_squish() function from the stringr package cleans such text by removing extraneous whitespace.
In this example, you will apply str_squish() to all character columns in the msleep dataset:
#> # A tibble: 83 × 11
#> name genus vore order conservation sleep_total sleep_rem sleep_cycle awake
#> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Cheet… Acin… carni Carn… lc 12.1 NA NA 11.9
#> 2 Owl m… Aotus omni Prim… <NA> 17 1.8 NA 7
#> 3 Mount… Aplo… herbi Rode… nt 14.4 2.4 NA 9.6
#> 4 Great… Blar… omni Sori… lc 14.9 2.3 0.133 9.1
#> 5 Cow Bos herbi Arti… domesticated 4 0.7 0.667 20
#> 6 Three… Brad… herbi Pilo… <NA> 14.4 2.2 0.767 9.6
#> 7 North… Call… carni Carn… vu 8.7 1.4 0.383 15.3
#> 8 Vespe… Calo… <NA> Rode… <NA> 7 NA NA 17
#> 9 Dog Canis carni Carn… domesticated 10.1 2.9 0.333 13.9
#> 10 Roe d… Capr… herbi Arti… lc 3 NA NA 21
#> # ℹ 73 more rows
#> # ℹ 2 more variables: brainwt <dbl>, bodywt <dbl>where(is.character): Selects all character columns.str_squish: Removes extra whitespace from the text in these columns.
For example, if a column contains the value " African elephant ", it will be cleaned up to "African elephant".
5.7.4 filter() – Selecting Rows Based on Conditions
The filter() function allows you to keep rows that meet specific criteria. Think of it as a way to zoom in on the part of your dataset that matters most.
Key Points:
filter()takes a logical condition (e.g.,gender == "female") and keeps rows where the condition isTRUE.-
You can use
filter()to subset numeric variables based on their values. Common comparison operators include:>(greater than)>=(greater than or equal to)<(less than)<=(less than or equal to)==(equal to)!=(not equal to)
For details on these operators, see Chapter 1.6.4.
-
To filter rows based on multiple conditions, you can use logical operators:
&or,(AND): Ensures all conditions are true.|(OR): Keeps rows where at least one condition is true.!(NOT): Negates a condition, keeping rows where it is false.
5.7.4.1 Basic row filters
Let’s find all animals that sleep more than 10 hours:
#> # A tibble: 44 × 2
#> name sleep_total
#> <chr> <dbl>
#> 1 Cheetah 12.1
#> 2 Owl monkey 17
#> 3 Mountain beaver 14.4
#> 4 Greater short-tailed shrew 14.9
#> 5 Three-toed sloth 14.4
#> 6 Dog 10.1
#> 7 Chinchilla 12.5
#> 8 Star-nosed mole 10.3
#> 9 Long-nosed armadillo 17.4
#> 10 North American Opossum 18
#> # ℹ 34 more rowsTo select a range of values, you can use two logical conditions. For example, to filter all animals with a total sleep time between 9 and 16 hours, you could use:
#> # A tibble: 46 × 2
#> name sleep_total
#> <chr> <dbl>
#> 1 Cheetah 12.1
#> 2 Mountain beaver 14.4
#> 3 Greater short-tailed shrew 14.9
#> 4 Three-toed sloth 14.4
#> 5 Dog 10.1
#> 6 Guinea pig 9.4
#> 7 Grivet 10
#> 8 Chinchilla 12.5
#> 9 Star-nosed mole 10.3
#> 10 Lesser short-tailed shrew 9.1
#> # ℹ 36 more rowsHowever, there is a more concise way to achieve the same result using the between() function6:
#> # A tibble: 46 × 2
#> name sleep_total
#> <chr> <dbl>
#> 1 Cheetah 12.1
#> 2 Mountain beaver 14.4
#> 3 Greater short-tailed shrew 14.9
#> 4 Three-toed sloth 14.4
#> 5 Dog 10.1
#> 6 Guinea pig 9.4
#> 7 Grivet 10
#> 8 Chinchilla 12.5
#> 9 Star-nosed mole 10.3
#> 10 Lesser short-tailed shrew 9.1
#> # ℹ 36 more rows5.7.4.2 Filtering Based on Exact Character Variable Matches
When working with character variables, you can filter rows based on exact matches, exclusions, or membership in specific groups. Below are practical examples of how to handle these scenarios effectively:
1. Select Rows with an Exact Match
To filter a specific group of animals, use the == comparison operator. For example, to select only carnivores:
#> # A tibble: 19 × 3
#> name vore sleep_total
#> <chr> <chr> <dbl>
#> 1 Cheetah carni 12.1
#> 2 Northern fur seal carni 8.7
#> 3 Dog carni 10.1
#> 4 Long-nosed armadillo carni 17.4
#> 5 Domestic cat carni 12.5
#> 6 Pilot whale carni 2.7
#> 7 Gray seal carni 6.2
#> 8 Thick-tailed opposum carni 19.4
#> 9 Slow loris carni 11
#> 10 Northern grasshopper mouse carni 14.5
#> 11 Tiger carni 15.8
#> 12 Jaguar carni 10.4
#> 13 Lion carni 13.5
#> 14 Caspian seal carni 3.5
#> 15 Common porpoise carni 5.6
#> 16 Bottle-nosed dolphin carni 5.2
#> 17 Genet carni 6.3
#> 18 Arctic fox carni 12.5
#> 19 Red fox carni 9.82. Exclude Rows Using !=
To exclude rows with a specific value, use the != operator. For example, to exclude omnivores:
#> # A tibble: 56 × 3
#> name vore sleep_total
#> <chr> <chr> <dbl>
#> 1 Cheetah carni 12.1
#> 2 Mountain beaver herbi 14.4
#> 3 Cow herbi 4
#> 4 Three-toed sloth herbi 14.4
#> 5 Northern fur seal carni 8.7
#> 6 Dog carni 10.1
#> 7 Roe deer herbi 3
#> 8 Goat herbi 5.3
#> 9 Guinea pig herbi 9.4
#> 10 Chinchilla herbi 12.5
#> # ℹ 46 more rows3. Filter for Multiple Values Using %in%
If you want to filter rows where a variable matches one of multiple values, use the %in% operator. For example, to select animals belonging to the orders Primates or Rodentia:
#> # A tibble: 34 × 3
#> name sleep_total order
#> <chr> <dbl> <chr>
#> 1 Owl monkey 17 Primates
#> 2 Mountain beaver 14.4 Rodentia
#> 3 Vesper mouse 7 Rodentia
#> 4 Guinea pig 9.4 Rodentia
#> 5 Grivet 10 Primates
#> 6 Chinchilla 12.5 Rodentia
#> 7 African giant pouched rat 8.3 Rodentia
#> 8 Patas monkey 10.9 Primates
#> 9 Western american chipmunk 14.9 Rodentia
#> 10 Galago 9.8 Primates
#> # ℹ 24 more rows4. Exclude Multiple Groups Using !%in%
To exclude rows belonging to specific groups, negate the %in% operator by using ! at the beginning of your filter. For example, to exclude animals in the orders Rodentia, Carnivora, and Primates:
msleep |>
select(name, order, sleep_total) |>
filter(!order %in% c("Rodentia", "Carnivora", "Primates"))#> # A tibble: 37 × 3
#> name order sleep_total
#> <chr> <chr> <dbl>
#> 1 Greater short-tailed shrew Soricomorpha 14.9
#> 2 Cow Artiodactyla 4
#> 3 Three-toed sloth Pilosa 14.4
#> 4 Roe deer Artiodactyla 3
#> 5 Goat Artiodactyla 5.3
#> 6 Star-nosed mole Soricomorpha 10.3
#> 7 Lesser short-tailed shrew Soricomorpha 9.1
#> 8 Long-nosed armadillo Cingulata 17.4
#> 9 Tree hyrax Hyracoidea 5.3
#> 10 North American Opossum Didelphimorphia 18
#> # ℹ 27 more rowsIn R, you must place the negation operator (!) before the variable you want to filter with when using the %in% operator. For example, instead of writing:
filter(order !%in% c("Rodentia", "Carnivora", "Primates"))—which is invalid—write it as:
This ensures that the filter correctly excludes the specified groups.
5.7.4.3 Filtering Rows Based on Regular Expressions
The filtering methods discussed earlier work well when you are matching the entire content of a variable. However, in many cases, you may need to filter rows based on partial matches within a string. To achieve this, you can use functions that evaluate regular expressions and return Boolean values (TRUE or FALSE). Rows where the condition is TRUE will be retained.
There are two ways to do this:
-
grepl()(from base R):Checks if a pattern exists in a string and returns a logical vector.
Often combined with
tolower()to make matching case-insensitive.
-
str_detect()(from thestringrpackage):A more intuitive function for detecting patterns in strings.
Often combined with
str_to_lower()to make matching case-insensitive.Part of the
tidyverse, making it consistent withdplyrworkflows.
R is case-sensitive by default. For instance:
- Using
filter(str_detect(name, pattern = "mouse"))would exclude rows with"Mouse"because of the difference in case.
To avoid missing such matches, it’s a good practice to convert the text to lowercase (or uppercase) using str_to_lower() (or str_to_upper()) before performing the match.
In the following example, we filter rows where the name column contains the substring “mouse,” regardless of case:
msleep |>
select(name, sleep_total) |>
filter(str_detect(str_to_lower(name), pattern = "mouse"))#> # A tibble: 5 × 2
#> name sleep_total
#> <chr> <dbl>
#> 1 Vesper mouse 7
#> 2 House mouse 12.5
#> 3 Northern grasshopper mouse 14.5
#> 4 Deer mouse 11.5
#> 5 African striped mouse 8.75.7.4.4 Filtering with Multiple Conditions
The filter() function not only allows filtering based on single conditions but also supports combining multiple conditions using logical operators. These operators—AND, OR, NOT, and XOR—as summarized in Table 5.1, provide the flexibility to create complex filtering logic tailored to your data analysis needs.
| Example | Operator | Description | Example Usage |
|---|---|---|---|
| 1 |
, or & (AND) |
Both conditions must be true for a row to be returned. |
filter(condition1, condition2) or filter(condition1 & condition2)
|
| 2 |
| (OR) |
At least one condition must be true for a row to be included. | filter(condition1 |condition2) |
| 3 |
! (NOT) |
The condition must be false for a row to be included. | filter(!condition1) |
| 4 | xor() |
Only one condition must be true, and not both. | filter(xor(condition1, condition2)) |
Example 1:
Filter Animals That Are Carnivores and Sleep More Than 10 Hours
msleep |>
filter(vore == "carni", sleep_total > 10)#> # A tibble: 11 × 11
#> name genus vore order conservation sleep_total sleep_rem sleep_cycle awake
#> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Cheet… Acin… carni Carn… lc 12.1 NA NA 11.9
#> 2 Dog Canis carni Carn… domesticated 10.1 2.9 0.333 13.9
#> 3 Long-… Dasy… carni Cing… lc 17.4 3.1 0.383 6.6
#> 4 Domes… Felis carni Carn… domesticated 12.5 3.2 0.417 11.5
#> 5 Thick… Lutr… carni Dide… lc 19.4 6.6 NA 4.6
#> 6 Slow … Nyct… carni Prim… <NA> 11 NA NA 13
#> 7 North… Onyc… carni Rode… lc 14.5 NA NA 9.5
#> 8 Tiger Pant… carni Carn… en 15.8 NA NA 8.2
#> 9 Jaguar Pant… carni Carn… nt 10.4 NA NA 13.6
#> 10 Lion Pant… carni Carn… vu 13.5 NA NA 10.5
#> 11 Arcti… Vulp… carni Carn… <NA> 12.5 NA NA 11.5
#> # ℹ 2 more variables: brainwt <dbl>, bodywt <dbl>Example 2:
Filter Animals That Are Omnivores or Sleep Less Than 8 Hours
msleep |>
filter(vore == "omni" | sleep_total < 8)#> # A tibble: 41 × 11
#> name genus vore order conservation sleep_total sleep_rem sleep_cycle awake
#> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Owl m… Aotus omni Prim… <NA> 17 1.8 NA 7
#> 2 Great… Blar… omni Sori… lc 14.9 2.3 0.133 9.1
#> 3 Cow Bos herbi Arti… domesticated 4 0.7 0.667 20
#> 4 Vespe… Calo… <NA> Rode… <NA> 7 NA NA 17
#> 5 Roe d… Capr… herbi Arti… lc 3 NA NA 21
#> 6 Goat Capri herbi Arti… lc 5.3 0.6 NA 18.7
#> 7 Grivet Cerc… omni Prim… lc 10 0.7 NA 14
#> 8 Star-… Cond… omni Sori… lc 10.3 2.2 NA 13.7
#> 9 Afric… Cric… omni Rode… <NA> 8.3 2 NA 15.7
#> 10 Lesse… Cryp… omni Sori… lc 9.1 1.4 0.15 14.9
#> # ℹ 31 more rows
#> # ℹ 2 more variables: brainwt <dbl>, bodywt <dbl>Example 3:
Filter Animals That Are Not Omnivores and Sleep More Than 8 Hours
msleep |>
filter(!vore == "herbi", sleep_total > 8)#> # A tibble: 37 × 11
#> name genus vore order conservation sleep_total sleep_rem sleep_cycle awake
#> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Cheet… Acin… carni Carn… lc 12.1 NA NA 11.9
#> 2 Owl m… Aotus omni Prim… <NA> 17 1.8 NA 7
#> 3 Great… Blar… omni Sori… lc 14.9 2.3 0.133 9.1
#> 4 North… Call… carni Carn… vu 8.7 1.4 0.383 15.3
#> 5 Dog Canis carni Carn… domesticated 10.1 2.9 0.333 13.9
#> 6 Grivet Cerc… omni Prim… lc 10 0.7 NA 14
#> 7 Star-… Cond… omni Sori… lc 10.3 2.2 NA 13.7
#> 8 Afric… Cric… omni Rode… <NA> 8.3 2 NA 15.7
#> 9 Lesse… Cryp… omni Sori… lc 9.1 1.4 0.15 14.9
#> 10 Long-… Dasy… carni Cing… lc 17.4 3.1 0.383 6.6
#> # ℹ 27 more rows
#> # ℹ 2 more variables: brainwt <dbl>, bodywt <dbl>Example 4:
Filter Animals That Are Either Carnivores or Sleep More Than 16 Hours, But Not Both
#> # A tibble: 23 × 11
#> name genus vore order conservation sleep_total sleep_rem sleep_cycle awake
#> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Cheet… Acin… carni Carn… lc 12.1 NA NA 11.9
#> 2 Owl m… Aotus omni Prim… <NA> 17 1.8 NA 7
#> 3 North… Call… carni Carn… vu 8.7 1.4 0.383 15.3
#> 4 Dog Canis carni Carn… domesticated 10.1 2.9 0.333 13.9
#> 5 North… Dide… omni Dide… lc 18 4.9 0.333 6
#> 6 Big b… Epte… inse… Chir… lc 19.7 3.9 0.117 4.3
#> 7 Domes… Felis carni Carn… domesticated 12.5 3.2 0.417 11.5
#> 8 Pilot… Glob… carni Ceta… cd 2.7 0.1 NA 21.4
#> 9 Gray … Hali… carni Carn… lc 6.2 1.5 NA 17.8
#> 10 Littl… Myot… inse… Chir… <NA> 19.9 2 0.2 4.1
#> # ℹ 13 more rows
#> # ℹ 2 more variables: brainwt <dbl>, bodywt <dbl>Order Matters: R evaluates conditions left to right in a
filter()statement.Combine Freely: You can mix
AND,OR, andNOTto form highly specific filters.Readability: Use parentheses for complex conditions to make them easier to understand.
For complex conditions combining AND (&), OR (|), and NOT (!), the order of evaluation is determined by standard precedence rules unless overridden with parentheses.
Precedence Order:
NOT (
!) is evaluated first.AND (
&or,) is evaluated second.OR (
|) is evaluated last.
Example 5:
Filter animals that are either herbivores or omnivores and have a total sleep time of more than 10 hours.
msleep |>
filter((vore == "herbi" | vore == "omni") & sleep_total > 10)#> # A tibble: 25 × 11
#> name genus vore order conservation sleep_total sleep_rem sleep_cycle awake
#> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Owl m… Aotus omni Prim… <NA> 17 1.8 NA 7
#> 2 Mount… Aplo… herbi Rode… nt 14.4 2.4 NA 9.6
#> 3 Great… Blar… omni Sori… lc 14.9 2.3 0.133 9.1
#> 4 Three… Brad… herbi Pilo… <NA> 14.4 2.2 0.767 9.6
#> 5 Chinc… Chin… herbi Rode… domesticated 12.5 1.5 0.117 11.5
#> 6 Star-… Cond… omni Sori… lc 10.3 2.2 NA 13.7
#> 7 North… Dide… omni Dide… lc 18 4.9 0.333 6
#> 8 Europ… Erin… omni Erin… lc 10.1 3.5 0.283 13.9
#> 9 Patas… Eryt… omni Prim… lc 10.9 1.1 NA 13.1
#> 10 Weste… Euta… herbi Rode… <NA> 14.9 NA NA 9.1
#> # ℹ 15 more rows
#> # ℹ 2 more variables: brainwt <dbl>, bodywt <dbl>If the parentheses were omitted, the conditions could be misinterpreted, leading to unintended results.
5.7.4.5 Filtering Out Empty Rows
Missing values (NA) in datasets can disrupt analysis, so it is often necessary to remove rows with missing values in specific columns. This can be done using the is.na() function within a filter() statement.
1. Select Rows Where a Column Has NA Values
To display rows where the conservation column has missing values (NA):
#> # A tibble: 29 × 5
#> name conservation sleep_total sleep_rem sleep_cycle
#> <chr> <chr> <dbl> <dbl> <dbl>
#> 1 "Owl monkey" <NA> 17 1.8 NA
#> 2 "Three-toed sloth" <NA> 14.4 2.2 0.767
#> 3 "Vesper mouse" <NA> 7 NA NA
#> 4 "African giant pouched rat" <NA> 8.3 2 NA
#> 5 "Western american chipmunk" <NA> 14.9 NA NA
#> 6 "Galago" <NA> 9.8 1.1 0.55
#> 7 "Human" <NA> 8 1.9 1.5
#> 8 "Macaque" <NA> 10.1 1.2 0.75
#> 9 "Vole " <NA> 12.8 NA NA
#> 10 "Little brown bat" <NA> 19.9 2 0.2
#> # ℹ 19 more rows2. Remove Rows Where a Column Has NA Values
To exclude rows where the conservation column is missing, negate the is.na() function using !:
#> # A tibble: 54 × 5
#> name conservation sleep_total sleep_rem sleep_cycle
#> <chr> <chr> <dbl> <dbl> <dbl>
#> 1 Cheetah lc 12.1 NA NA
#> 2 Mountain beaver nt 14.4 2.4 NA
#> 3 Greater short-tailed shrew lc 14.9 2.3 0.133
#> 4 Cow domesticated 4 0.7 0.667
#> 5 Northern fur seal vu 8.7 1.4 0.383
#> 6 Dog domesticated 10.1 2.9 0.333
#> 7 Roe deer lc 3 NA NA
#> 8 Goat lc 5.3 0.6 NA
#> 9 Guinea pig domesticated 9.4 0.8 0.217
#> 10 Grivet lc 10 0.7 NA
#> # ℹ 44 more rows3. Filtering Multiple Columns for Missing Values
To remove rows with missing values across multiple columns, you can combine multiple is.na() checks:
msleep |>
select(name, conservation, sleep_total) |>
filter(!is.na(conservation), !is.na(sleep_total))#> # A tibble: 54 × 3
#> name conservation sleep_total
#> <chr> <chr> <dbl>
#> 1 Cheetah lc 12.1
#> 2 Mountain beaver nt 14.4
#> 3 Greater short-tailed shrew lc 14.9
#> 4 Cow domesticated 4
#> 5 Northern fur seal vu 8.7
#> 6 Dog domesticated 10.1
#> 7 Roe deer lc 3
#> 8 Goat lc 5.3
#> 9 Guinea pig domesticated 9.4
#> 10 Grivet lc 10
#> # ℹ 44 more rows4. Using if_any() or if_all() for Cleaner Filtering
The if_any() and if_all() functions from dplyr offer a more concise way to handle missing values across multiple columns:
- a. Remove Rows with
NAin Any Selected Column
This keeps rows where at least one of the specified columns (conservation or sleep_total) is not NA:
msleep |>
select(name, conservation, sleep_total) |>
filter(if_any(c(conservation, sleep_total), ~ !is.na(.)))#> # A tibble: 83 × 3
#> name conservation sleep_total
#> <chr> <chr> <dbl>
#> 1 Cheetah lc 12.1
#> 2 Owl monkey <NA> 17
#> 3 Mountain beaver nt 14.4
#> 4 Greater short-tailed shrew lc 14.9
#> 5 Cow domesticated 4
#> 6 Three-toed sloth <NA> 14.4
#> 7 Northern fur seal vu 8.7
#> 8 Vesper mouse <NA> 7
#> 9 Dog domesticated 10.1
#> 10 Roe deer lc 3
#> # ℹ 73 more rows- b. Remove Rows with
NAin All Selected Columns
This keeps rows where all the specified columns are not NA:
msleep |>
select(name, conservation, sleep_total) |>
filter(if_all(c(conservation, sleep_total), ~ !is.na(.)))#> # A tibble: 54 × 3
#> name conservation sleep_total
#> <chr> <chr> <dbl>
#> 1 Cheetah lc 12.1
#> 2 Mountain beaver nt 14.4
#> 3 Greater short-tailed shrew lc 14.9
#> 4 Cow domesticated 4
#> 5 Northern fur seal vu 8.7
#> 6 Dog domesticated 10.1
#> 7 Roe deer lc 3
#> 8 Goat lc 5.3
#> 9 Guinea pig domesticated 9.4
#> 10 Grivet lc 10
#> # ℹ 44 more rowsBy filtering out rows with missing values, you ensure that your analysis is accurate, clean, and focused on complete data.
5.7.5 arrange() – Reordering Rows
The arrange() function lets you sort rows in a dataset based on the values in one or more columns. Sorting can be ascending or descending.
Key Points:
By default, rows are sorted in ascending order.
To sort in descending order, use the
desc()function
Example 1: Sort by Total Sleep Time (Ascending)
Suppose you want to focus on species belonging to the orders Rodentia, Carnivora, and Primates. After filtering these groups, you can arrange the rows by their total sleep time (sleep_total) to identify species that sleep the least.
# Total sleep in ascending order
msleep |>
select(name, order, sleep_total) |>
filter(order %in% c("Rodentia", "Carnivora", "Primates")) |>
arrange(sleep_total)#> # A tibble: 46 × 3
#> name order sleep_total
#> <chr> <chr> <dbl>
#> 1 Caspian seal Carnivora 3.5
#> 2 Gray seal Carnivora 6.2
#> 3 Genet Carnivora 6.3
#> 4 Vesper mouse Rodentia 7
#> 5 Degu Rodentia 7.7
#> 6 Human Primates 8
#> 7 African giant pouched rat Rodentia 8.3
#> 8 Northern fur seal Carnivora 8.7
#> 9 African striped mouse Rodentia 8.7
#> 10 Guinea pig Rodentia 9.4
#> # ℹ 36 more rowsExample 2: Sort by Total Sleep Time (Descending)
To identify the species that sleep the most, you can sort the same filtered dataset in descending order of sleep_total:
# Total sleep in descending order
msleep |>
select(name, order, sleep_total) |>
filter(order %in% c("Rodentia", "Carnivora", "Primates")) |>
arrange(desc(sleep_total))#> # A tibble: 46 × 3
#> name order sleep_total
#> <chr> <chr> <dbl>
#> 1 Owl monkey Primates 17
#> 2 Arctic ground squirrel Rodentia 16.6
#> 3 Golden-mantled ground squirrel Rodentia 15.9
#> 4 Tiger Carnivora 15.8
#> 5 Eastern american chipmunk Rodentia 15.8
#> 6 Western american chipmunk Rodentia 14.9
#> 7 Round-tailed muskrat Rodentia 14.6
#> 8 Northern grasshopper mouse Rodentia 14.5
#> 9 Mountain beaver Rodentia 14.4
#> 10 Golden hamster Rodentia 14.3
#> # ℹ 36 more rows
5.7.6 slice() – Selecting Rows by Position
The slice() function is a simple and efficient way to select rows based on their numerical position in your dataset7. Unlike functions that filter rows based on the content of columns, slice() simply picks rows by their numeric index. There are several helpful variants of this function that you might find useful:
-
slice_head(): Selects the first n rows. -
slice_tail(): Selects the last n rows. -
slice_sample(): Randomly selects a specified number of rows. -
slice_min()andslice_max(): Select rows with the minimum or maximum values of a given variable.
Example 1: Select the First 5 Rows
Imagine you wish to quickly inspect the top entries from species belonging to the orders Rodentia, Carnivora, and Primates. With slice_head(), you can easily view the first 5 rows after filtering and selecting the relevant columns.
# Selecting the first 5 rows
msleep |>
select(name, order, sleep_total) |>
filter(order %in% c("Rodentia", "Carnivora", "Primates")) |>
slice_head(n = 5)#> # A tibble: 5 × 3
#> name order sleep_total
#> <chr> <chr> <dbl>
#> 1 Cheetah Carnivora 12.1
#> 2 Owl monkey Primates 17
#> 3 Mountain beaver Rodentia 14.4
#> 4 Northern fur seal Carnivora 8.7
#> 5 Vesper mouse Rodentia 7Example 2: Select the Last 5 Rows
Similarly, if you are interested in looking at the bottom entries of the same group of species, you can use slice_tail() to view the last 5 rows.
# Selecting the last 5 rows
msleep |>
select(name, order, sleep_total) |>
filter(order %in% c("Rodentia", "Carnivora", "Primates")) |>
slice_tail(n = 5)#> # A tibble: 5 × 3
#> name order sleep_total
#> <chr> <chr> <dbl>
#> 1 Golden-mantled ground squirrel Rodentia 15.9
#> 2 Eastern american chipmunk Rodentia 15.8
#> 3 Genet Carnivora 6.3
#> 4 Arctic fox Carnivora 12.5
#> 5 Red fox Carnivora 9.8Both slice_head() and slice_tail() are very useful when you want to quickly check the beginning or end of a filtered dataset, especially when dealing with large datasets.
Example 3: Identify Top 5 Animals by Brain-to-Body Weight Ratio Using slice_max()
Suppose you want to identify the top five animals with the highest brain-to-body weight ratio among those that weigh more than 5 kg and have available brain weight data. First, filter the dataset to include only animals meeting these criteria. Then, calculate the ratio by dividing brainwt by bodywt. Finally, the slice_max() function will help you extract the five animals with the highest ratios.
msleep |>
filter(bodywt > 5, !is.na(brainwt)) |>
mutate(brain_to_body_ratio = brainwt / bodywt) |>
slice_max(brain_to_body_ratio, n = 5)#> # A tibble: 5 × 12
#> name genus vore order conservation sleep_total sleep_rem sleep_cycle awake
#> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Macaque Maca… omni Prim… <NA> 10.1 1.2 0.75 13.9
#> 2 Human Homo omni Prim… <NA> 8 1.9 1.5 16
#> 3 Patas … Eryt… omni Prim… lc 10.9 1.1 NA 13.1
#> 4 Chimpa… Pan omni Prim… <NA> 9.7 1.4 1.42 14.3
#> 5 Baboon Papio omni Prim… <NA> 9.4 1 0.667 14.6
#> # ℹ 3 more variables: brainwt <dbl>, bodywt <dbl>, brain_to_body_ratio <dbl>Example 4: Identify Bottom 3 Species by REM Sleep Using slice_min()
Suppose you want to find out which species have the least amount of REM sleep among those in the orders Rodentia, Carnivora, and Primates. The slice_min() function will help you extract the three species with the minimum REM sleep values.
msleep |>
select(name, order, sleep_rem) |>
filter(order %in% c("Rodentia", "Carnivora", "Primates")) |>
slice_min(sleep_rem, n = 3)#> # A tibble: 3 × 3
#> name order sleep_rem
#> <chr> <chr> <dbl>
#> 1 Caspian seal Carnivora 0.4
#> 2 Grivet Primates 0.7
#> 3 Guinea pig Rodentia 0.8When you use slice_min() or slice_max(), the rows returned are already sorted by the specified variable (ascending for slice_min() and descending for slice_max()). You only need to use arrange() if you want to apply a different sorting order than the default.
5.7.7 summarise() – Aggregating Data
The summarise() function is used to create summary statistics by collapsing data into single values, such as calculating the minimum, maximum, mean, median, standard deviation, sum, or count for specific variables.
To use summarise(), define a new column name, followed by the = sign and the summary calculation:new_column = function(variable). You can include multiple summary functions within a single summarise() statement.
Key Points:
Use it to compute one or more summary statistics.
The functions
summarise()andsummarize()are interchangeableIt is often used with
group_by()to generate summaries by groups within the dataset.
Example: Summarising the Entire Dataset
To calculate the total number of animals, the average sleep time, and the maximum sleep time across all species in the msleep dataset:
#> # A tibble: 1 × 3
#> n average maximum
#> <int> <dbl> <dbl>
#> 1 83 10.4 19.9The summarise() function works with various aggregate functions, including:
n(): Number of observations.n_distinct(var): Number of unique values in a variable.Arithmetic functions:
sum(var),max(var),min(var).Statistical functions:
mean(var),median(var),sd(var),IQR(var).
In most cases, we don’t just want to summarise the whole data table, but we want to get summaries by a group.
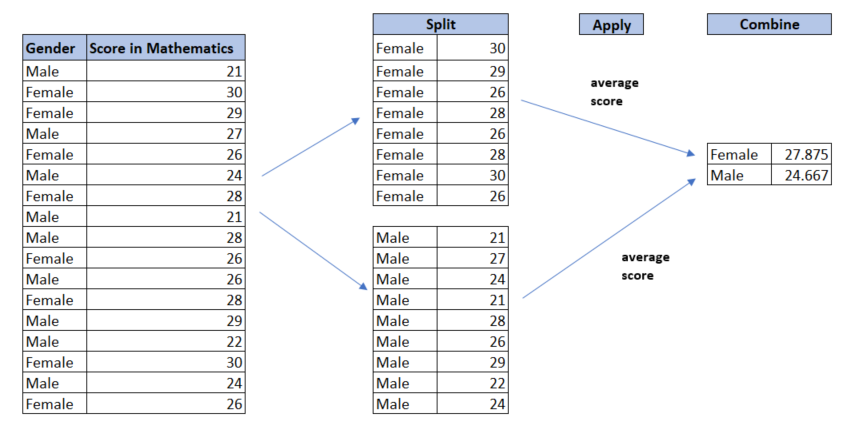
5.7.8 group_by() – Working with Groups
On its own, the group_by() function doesn’t perform any operation. However, when combined with functions like summarise() or mutate(), it becomes a powerful tool for splitting data into groups and applying operations to each group separately.
Key Points:
Groups can be based on one or multiple variables.
After grouping, any operation is applied independently to each group.
The results are combined into a single data frame.
Figure 5.5 below illustrates the group by strategy:
Example 2: Summarising by Groups
To generate summaries for specific groups, combine summarise() with group_by():
msleep |>
group_by(order) |>
summarise(
n = n(),
average_sleep = mean(sleep_total, na.rm = TRUE),
maximum_sleep = max(sleep_total, na.rm = TRUE)
)#> # A tibble: 19 × 4
#> order n average_sleep maximum_sleep
#> <chr> <int> <dbl> <dbl>
#> 1 Afrosoricida 1 15.6 15.6
#> 2 Artiodactyla 6 4.52 9.1
#> 3 Carnivora 12 10.1 15.8
#> 4 Cetacea 3 4.5 5.6
#> 5 Chiroptera 2 19.8 19.9
#> 6 Cingulata 2 17.8 18.1
#> 7 Didelphimorphia 2 18.7 19.4
#> 8 Diprotodontia 2 12.4 13.7
#> 9 Erinaceomorpha 2 10.2 10.3
#> 10 Hyracoidea 3 5.67 6.3
#> 11 Lagomorpha 1 8.4 8.4
#> 12 Monotremata 1 8.6 8.6
#> 13 Perissodactyla 3 3.47 4.4
#> 14 Pilosa 1 14.4 14.4
#> 15 Primates 12 10.5 17
#> 16 Proboscidea 2 3.6 3.9
#> 17 Rodentia 22 12.5 16.6
#> 18 Scandentia 1 8.9 8.9
#> 19 Soricomorpha 5 11.1 14.9In this example:
n: Number of species in each group.average_sleep: Average sleep time for each group, ignoringNAvalues (na.rm = TRUE).maximum_sleep: Maximum sleep time for each group, ignoringNAvalues.
Using summarise() with or without group_by() helps you aggregate and summarize your data efficiently, making it easier to extract meaningful insights from your dataset.
5.7.9 Combining All the Verbs:
Let’s tackle a realistic problem by chaining the verbs you’ve learned together. For this example, we’ll use the penguins dataset (from the palmerpenguins package) to answer a specific question:
Task:
Identify the top 5 penguins on Dream Island with flipper lengths exceeding 200 mm. Calculate their bill aspect ratio (bill length ÷ bill depth) and display the results sorted by this ratio.
# Load the dataset
penguins <- palmerpenguins::penguins
# Perform the analysis
penguins |>
filter(island == "Dream" & flipper_length_mm > 200) |>
mutate(bill_aspect_ratio = bill_length_mm / bill_depth_mm) |>
slice_max(bill_aspect_ratio, n = 5) |>
select(species, flipper_length_mm, bill_aspect_ratio)#> # A tibble: 5 × 3
#> species flipper_length_mm bill_aspect_ratio
#> <fct> <int> <dbl>
#> 1 Chinstrap 201 2.87
#> 2 Chinstrap 207 2.82
#> 3 Chinstrap 201 2.75
#> 4 Chinstrap 203 2.71
#> 5 Chinstrap 201 2.71This code performs the following steps:
Filter: Retains only penguins located on Dream Island with a flipper length greater than 200 mm.
Mutate: Calculates the
bill_aspect_ratioby dividingbill_length_mmbybill_depth_mm.Slice Max: Selects the top 5 penguins with the highest bill aspect ratios.
Select: Displays only the columns for species, flipper length, and bill aspect ratio.
5.7.10 Exercise 5.2.1: Top 5 Carnivorous Animals
Now, let’s extend your skills with a new challenge. Using the msleep dataset, identify the top 5 carnivorous animals that sleep the most and calculate their sleep-to-weight ratio to understand how sleep duration scales with body size.
Task: Complete the following code by replacing the placeholders (...) with the correct values:
msleep |>
filter(vore == ...) |>
mutate(sleep_to_weight = ... / ...) |>
select(name, sleep_total, sleep_to_weight) |>
slice_max(sleep_total, n = ---)Replace
...with the appropriate filtering criteria and calculations.Ensure the final code filters for carnivores, calculates the
sleep_to-weightratio, and returns the top 5 animals that sleep the most.
5.7.11 Exploring More Functions in dplyr
The dplyr package offers a wealth of functions to simplify data manipulation, enabling you to efficiently clean, transform, and analyze data. In addition to the core verbs (filter(), select(), mutate(), arrange(), summarize()), there are other incredibly useful functions such as:
rename(): Rename columns in your dataset.distinct(): Extract unique rows or values.count(): Count occurrences of unique values in a variable.relocate(): Reorder or reposition columns for better organization.
| Function | Purpose | Example Usage |
|---|---|---|
rename() |
Rename columns to more meaningful names | rename(new_name = old_name) |
distinct() |
Find unique rows or specific values | distinct(column1, column2) |
count() |
Count the frequency of unique values | count(column_name) |
relocate() |
Reorder columns for better organization | relocate(column_name, .before = another_column) |
Let’s explore each of the functions in Table 5.2 in detail using the msleep dataset from ggplot2 and the penguins dataset from palmerpenguins.
5.7.11.1 rename() – Renaming Columns
The rename() function allows you to change column names to make them more meaningful or easier to work with. This is especially helpful when dealing with datasets that have poorly named columns.
Key Points:
- Syntax:
rename(new_name = old_name) - You can rename one or multiple columns at a time.
- The rest of the dataset remains unchanged.
Example 1: Renaming Columns in msleep
Rename the column name to animal_name and sleep_total to total_sleep:
msleep |>
rename(
animal_name = name,
total_sleep = sleep_total
)#> # A tibble: 83 × 11
#> animal_name genus vore order conservation total_sleep sleep_rem sleep_cycle
#> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl>
#> 1 Cheetah Acin… carni Carn… lc 12.1 NA NA
#> 2 Owl monkey Aotus omni Prim… <NA> 17 1.8 NA
#> 3 Mountain be… Aplo… herbi Rode… nt 14.4 2.4 NA
#> 4 Greater sho… Blar… omni Sori… lc 14.9 2.3 0.133
#> 5 Cow Bos herbi Arti… domesticated 4 0.7 0.667
#> 6 Three-toed … Brad… herbi Pilo… <NA> 14.4 2.2 0.767
#> 7 Northern fu… Call… carni Carn… vu 8.7 1.4 0.383
#> 8 Vesper mouse Calo… <NA> Rode… <NA> 7 NA NA
#> 9 Dog Canis carni Carn… domesticated 10.1 2.9 0.333
#> 10 Roe deer Capr… herbi Arti… lc 3 NA NA
#> # ℹ 73 more rows
#> # ℹ 3 more variables: awake <dbl>, brainwt <dbl>, bodywt <dbl>-
rename(animal_name = name)renames thenamecolumn toanimal_name. -
rename(total_sleep = sleep_total)renamessleep_totaltototal_sleep. - You can use this to make column names more descriptive.
Example 2:
Rename bill_length_mm to bill_length and flipper_length_mm to flipper_length in the penguins data:
penguins |>
rename(
bill_length = bill_length_mm,
flipper_length = flipper_length_mm
)#> # A tibble: 344 × 8
#> species island bill_length bill_depth_mm flipper_length body_mass_g sex
#> <fct> <fct> <dbl> <dbl> <int> <int> <fct>
#> 1 Adelie Torgersen 39.1 18.7 181 3750 male
#> 2 Adelie Torgersen 39.5 17.4 186 3800 female
#> 3 Adelie Torgersen 40.3 18 195 3250 female
#> 4 Adelie Torgersen NA NA NA NA <NA>
#> 5 Adelie Torgersen 36.7 19.3 193 3450 female
#> 6 Adelie Torgersen 39.3 20.6 190 3650 male
#> 7 Adelie Torgersen 38.9 17.8 181 3625 female
#> 8 Adelie Torgersen 39.2 19.6 195 4675 male
#> 9 Adelie Torgersen 34.1 18.1 193 3475 <NA>
#> 10 Adelie Torgersen 42 20.2 190 4250 <NA>
#> # ℹ 334 more rows
#> # ℹ 1 more variable: year <int>How might renaming confusing column names or removing duplicates before analysis contribute to more confident and accurate inferences?
5.7.11.2 distinct() – Extracting Unique Rows or Values
Duplicates in datasets can distort analyses and lead to inaccurate conclusions. Identifying and removing duplicates ensures clean data that accurately represents unique observations. The distinct() function is a simple yet powerful tool for finding unique rows or specific combinations of values in a dataset. It is particularly useful for removing duplicate rows or understanding unique categories in a variable.
Key Points:
Default Behavior:
By default,distinct()considers all columns to identify unique rows.Specific Variables:
You can specify one or more columns to extract unique values for specific variables.Keeping All Variables:
Adding.keep_all = TRUEwhile specifying columns ensures that all other variables are retained in the resulting dataset.-
Using
janitor::get_dupes():
Theget_dupes()function from the janitor package provides an easy way to identify duplicates. It returns:Full records where the specified variables have duplicates.
A column called
dupe_countshowing the number of rows sharing each duplicate combination.
Example 1: Counting Duplicates (iris Dataset)
To count the number of duplicate rows in a dataset, use the combination of duplicated() and sum() functions:
sum(duplicated(iris))#> [1] 1There is one duplicate record found in this dataset.
Example 2: Identifying and Removing Duplicates (iris Dataset)
To find duplicate records in the iris dataset
iris |> janitor::get_dupes()#> No variable names specified - using all columns.#> # A tibble: 2 × 6
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species dupe_count
#> <dbl> <dbl> <dbl> <dbl> <fct> <int>
#> 1 5.8 2.7 5.1 1.9 virginica 2
#> 2 5.8 2.7 5.1 1.9 virginica 2The output shows the duplicate records in the iris dataset. To remove these duplicates and retain only the unique rows, you can use the distinct() function as follows:
iris |> distinct()#> # A tibble: 149 × 5
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> <dbl> <dbl> <dbl> <dbl> <fct>
#> 1 5.1 3.5 1.4 0.2 setosa
#> 2 4.9 3 1.4 0.2 setosa
#> 3 4.7 3.2 1.3 0.2 setosa
#> 4 4.6 3.1 1.5 0.2 setosa
#> 5 5 3.6 1.4 0.2 setosa
#> 6 5.4 3.9 1.7 0.4 setosa
#> 7 4.6 3.4 1.4 0.3 setosa
#> 8 5 3.4 1.5 0.2 setosa
#> 9 4.4 2.9 1.4 0.2 setosa
#> 10 4.9 3.1 1.5 0.1 setosa
#> # ℹ 139 more rowsExample 3: Finding Unique Values in a Specific Column (msleep Dataset)
To extract unique values from the vore column in the msleep dataset (diet types):
msleep |>
distinct(vore)#> # A tibble: 5 × 1
#> vore
#> <chr>
#> 1 carni
#> 2 omni
#> 3 herbi
#> 4 <NA>
#> 5 insectiThe distinct(vore) function returns only the unique values in the vore column, helping you understand the categories in this variable.
Example 4: Keeping All Variables When Filtering for Uniqueness
To return unique rows based on the vore column while keeping all other variables:
msleep |>
distinct(vore, .keep_all = TRUE)#> # A tibble: 5 × 11
#> name genus vore order conservation sleep_total sleep_rem sleep_cycle awake
#> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Cheetah Acin… carni Carn… lc 12.1 NA NA 11.9
#> 2 Owl mo… Aotus omni Prim… <NA> 17 1.8 NA 7
#> 3 Mounta… Aplo… herbi Rode… nt 14.4 2.4 NA 9.6
#> 4 Vesper… Calo… <NA> Rode… <NA> 7 NA NA 17
#> 5 Big br… Epte… inse… Chir… lc 19.7 3.9 0.117 4.3
#> # ℹ 2 more variables: brainwt <dbl>, bodywt <dbl>Adding .keep_all = TRUE ensures that the uniqueness is determined by the vore column, but all other columns are retained in the output.
5.7.11.3 count() – Counting Occurrences
The count() function is a quick way to calculate the frequency of unique values in a column or combinations of columns. It is particularly useful for summarizing categorical variables.
Adding the sort = TRUE argument will automatically sort the results in descending order of frequency.
Key Points:
- By default,
count()returns the number of occurrences for each unique value. - Use
count(x, sort = TRUE)to sort the results, with the largest groups appearing at the top.
Example 1
Count the number of animals in each diet category (vore) in the msleep data:
msleep |>
count(vore, sort = TRUE)#> # A tibble: 5 × 2
#> vore n
#> <chr> <int>
#> 1 herbi 32
#> 2 omni 20
#> 3 carni 19
#> 4 <NA> 7
#> 5 insecti 5Explanation:
count(vore)calculates how many times each diet type (vore) appears.The output has two columns:
voreandn(the count).
Example 2
Count the number of penguins on each island in the penguins data:
penguins |>
count(island)#> # A tibble: 3 × 2
#> island n
#> <fct> <int>
#> 1 Biscoe 168
#> 2 Dream 124
#> 3 Torgersen 52
5.7.11.4 relocate() – Reordering Columns
The relocate() function allows you to rearrange columns for better readability or logical grouping using the same syntax as select() to make it easy to move blocks of columns at once. It doesn’t remove or modify columns, only changes their position.
Key Points:
Use
relocate(column_name, .before = ...)to move a column before a specific column.Use
relocate(column_name, .after = ...)to move a column after a specific column.
Example 1 : Reordering Columns in msleep
Move bodywt to appear after the sleep_total column:
msleep |>
relocate(bodywt, .after = sleep_total)#> # A tibble: 83 × 11
#> name genus vore order conservation sleep_total bodywt sleep_rem
#> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl>
#> 1 Cheetah Acin… carni Carn… lc 12.1 50 NA
#> 2 Owl monkey Aotus omni Prim… <NA> 17 0.48 1.8
#> 3 Mountain beaver Aplo… herbi Rode… nt 14.4 1.35 2.4
#> 4 Greater short-t… Blar… omni Sori… lc 14.9 0.019 2.3
#> 5 Cow Bos herbi Arti… domesticated 4 600 0.7
#> 6 Three-toed sloth Brad… herbi Pilo… <NA> 14.4 3.85 2.2
#> 7 Northern fur se… Call… carni Carn… vu 8.7 20.5 1.4
#> 8 Vesper mouse Calo… <NA> Rode… <NA> 7 0.045 NA
#> 9 Dog Canis carni Carn… domesticated 10.1 14 2.9
#> 10 Roe deer Capr… herbi Arti… lc 3 14.8 NA
#> # ℹ 73 more rows
#> # ℹ 3 more variables: sleep_cycle <dbl>, awake <dbl>, brainwt <dbl>Example 2:
You can also relocate variables based on their data type:
#> # A tibble: 344 × 8
#> species island sex bill_length_mm bill_depth_mm flipper_length_mm
#> <fct> <fct> <fct> <dbl> <dbl> <int>
#> 1 Adelie Torgersen male 39.1 18.7 181
#> 2 Adelie Torgersen female 39.5 17.4 186
#> 3 Adelie Torgersen female 40.3 18 195
#> 4 Adelie Torgersen <NA> NA NA NA
#> 5 Adelie Torgersen female 36.7 19.3 193
#> 6 Adelie Torgersen male 39.3 20.6 190
#> 7 Adelie Torgersen female 38.9 17.8 181
#> 8 Adelie Torgersen male 39.2 19.6 195
#> 9 Adelie Torgersen <NA> 34.1 18.1 193
#> 10 Adelie Torgersen <NA> 42 20.2 190
#> # ℹ 334 more rows
#> # ℹ 2 more variables: body_mass_g <int>, year <int>Combining These Functions
Let’s combine rename(), distinct(), count(), and relocate() to solve a real-world problem.
Example: Cleaning and Organizing msleep Data
Rename columns for clarity.
Remove duplicate rows.
Reorganize columns for better readability.
Count occurrences of diet types.
5.7.12 Practice Quiz 5.2
Question 1:
Which function would you use in dplyr to randomly select a specified number of rows from a dataset?
-
sample(n = 5)
-
slice_sample(n = 5)
-
filter_sample()
mutate_sample()
Question 2:
To calculate the average sleep_total for each vore category, which combination of functions is most appropriate?
group_by(vore) |> select(sleep_total) |> summarise(mean(sleep_total))select(vore, sleep_total) |> summarise(mean(sleep_total)) |> group_by(vore)group_by(vore) |> summarise(avg_sleep = mean(sleep_total, na.rm = TRUE))filter(vore) |> mutate(avg_sleep = mean(sleep_total))
Question 3:
To extract rows with the maximum value of a specified variable, which function is appropriate in dplyr?
Question 4:
Which dplyr function would you use if you want to create a new column called weight_ratio by dividing bodywt by mean_bodywt?
Question 5:
Suppose you need to identify the top 3 penguins with the highest bill aspect ratio from the penguins dataset after calculating it in a new column. Which of the following code snippets is the most concise and appropriate?
penguins |>
mutate(bill_aspect_ratio = bill_length_mm / bill_depth_mm) |>
arrange(desc(bill_aspect_ratio)) |>
head(3)penguins |>
mutate(bill_aspect_ratio = bill_length_mm / bill_depth_mm) |>
slice_max(bill_aspect_ratio, n = 3)Both a and b are equally concise and valid.
Neither a nor b is valid.
Question 6:
Given the following code, which is the correct equivalent using the pipe operator?
msleep |> select(name, sleep_total) |> filter(sleep_total > 8) |> arrange(sleep_total)msleep |> filter(sleep_total > 8) |> select(name, sleep_total) |> arrange(sleep_total)select(msleep, name, sleep_total) |> filter(sleep_total > 8) |> arrange(sleep_total)msleep |> arrange(sleep_total) |> filter(sleep_total > 8) |> select(name, sleep_total)
Question 7:
Which of the following correctly applies a log transformation to numeric columns only?
mutate_all(log)mutate(across(everything(), log))Question 8:
What does mutate(across(everything(), as.character)) do?
- Converts all character columns to numeric.
- Converts all columns in the dataset to character type.
- Applies a conditional transformation to numeric columns.
- Filters out non-character values.
Question 9:
To extract the rows with the minimum value of a specified variable, which dplyr function should you use?
Question 10:
If you want to reorder the rows of msleep by sleep_total in ascending order and then only show the top 5 rows, which code snippet is correct?
msleep |> arrange(sleep_total) |> head(5)msleep |> head(5) |> arrange(sleep_total)msleep |> summarise(sleep_total) |> head(5)msleep |> select(sleep_total) |> arrange(desc(sleep_total)) |> head(5)
5.7.13 Exercise 5.2.2: Analysing the Penguins Dataset
Let’s put your skills into practice with a modified penguins dataset. First, you’ll need to create a new RStudio project called Experiment 5.1.
-
Importing and Inspecting Data
Locate the
penguins.xlsxfile in ther-datadirectory. If you don’t already have the file, you can download it from Google Drive.Import the data into R.
Use
glimpse(penguins)to get an overview.How many rows and columns are there?
-
Filtering Data
How many penguins are from the
Biscoeisland?Extract data for penguins with a body mass greater than 4,500 grams.
-
Arranging Data
Arrange the data in descending order based on flipper length.
Find the top 5 penguins with the highest body mass.
-
Selecting and Mutating
Select only the columns
species,island, andsex.Remove the
sexcolumn from the dataset.-
Convert the flipper length from millimeters to meters and create a new column
flipper_length_mTo convert millimeters to meters, you simply divide the number of millimeters by 1,000. Here’s the conversion formula:
\[\text{flipper\_length\_m} = \frac{\text{flipper\_length\_mm}}{1000}\]
-
Create a new column
BMIcalculated as:\[\text{BMI} = \frac{\text{body\_mass\_g}}{{\text{flipper\_length\_m}}^2}\]
-
Summarizing and Grouping
Calculate the average body mass of all penguins.
Group the data by
speciesand find the average body mass for each species.
-
Combining Operations
- Filter penguins from the Dream island and summarize the average bill length for each species from this island.
5.7.14 Exercise 5.2.3: Data Analyst Candidate Assessment
In this exercise, you’ll work with a real dataset of medical insurance records similar to those of a well-known health insurance company in the country. We want to see how you clean, transform and analyse data in a practical, real-world context as a Data Analyst. Please follow the instructions below and document your process along the way.
Dataset Overview
You’re provided with a dataset containing medical insurance records for various individuals. Here’s what each column represents:
User ID: A unique identifier for each individual.
Gender: The individual’s gender (‘Male’ or ‘Female’).
Age: The age of the individual in years.
AgeGroup: The age bracket into which the individual falls.
Estimated Salary: An estimate of the individual’s annual salary.
Purchased: Indicates whether the individual has purchased medical insurance (‘purchased’ or ‘not-purchased’).
Data Import Instructions
Locate the
medical_insurance.xlsxfile in ther-datadirectory. If you don’t have it yet, you can download it from Google Drive.Import the dataset into R using the
readxlpackage.
Tasks
1. Data Transformation
Purchased Column Conversion:
Convert thePurchasedcolumn values to binary: use1for ‘purchased’ and0for ‘not-purchased’.-
Creating Salary Brackets:
Add a new column calledSalaryBracketbased on theEstimated Salary:-
Low: Salary < 30,000
-
Medium: Salary between 30,000 and 70,000
- High: Salary > 70,000
-
Low: Salary < 30,000
2. Analysis and Insights
-
Insurance Purchase Analysis:
Calculate and present:- The percentage of individuals who have purchased insurance, broken down by Gender.
- The percentage of individuals who have purchased insurance, broken down by AgeGroup.
Salary Bracket Purchase Rate:
Determine whichSalaryBracketshows the highest rate of insurance purchases.
Take your time to work through these tasks carefully. We’re looking forward to seeing how you apply your analytical skills to solve real-world data challenges. Good luck!
5.8 Experiment 5.3: Dealing with Missing Data
Missing data is common in real-world scenarios. Values may be missing because of measurement errors, data entry mistakes, or unavailability of certain information. It is crucial to detect and handle missing values properly, as they can bias results or cause errors in your analysis. R provides several functions to help you deal with missing data.
5.8.1 Recognising Missing Values
In R, missing values are represented by NA. Identifying these missing values is crucial for accurate data analysis. Here are some functions to check for missing data:
-
is.na(): Returns a logical vector indicating which elements areNA. -
anyNA(): Checks if there are anyNAvalues in an object. It returnsTRUEif there is at least oneNA, andFALSEotherwise.anyNA(x)#> [1] TRUE
Let’s apply the anyNA() function to a sample salary_data data frame:
salary_data <- data.frame(
Name = c("Alice", "Francisca", "Fatima", "David"),
Age = c(25, NA, 30, 35),
Salary = c(50000, 52000, NA, 55000)
)
salary_data#> Name Age Salary
#> 1 Alice 25 50000
#> 2 Francisca NA 52000
#> 3 Fatima 30 NA
#> 4 David 35 55000In this data frame, Francisca’s age and Fatima’s salary are missing. We can use anyNA() to check whether there are any missing values in the entire data frame:
anyNA(salary_data)#> [1] TRUESince the output is TRUE, we know there are missing values. We can also check specific columns for missing values. For example, let’s check the Age column:
anyNA(salary_data$Age)#> [1] TRUEAnd similarly, we can check the Name column:
anyNA(salary_data$Name)#> [1] FALSE-
complete.cases(): Identifies rows in a dataset that have no missing values (NA). It evaluates each row and checks whether it is “complete” (i.e., contains noNAvalues). It returns a logical vector where:TRUEindicates that a row has no missing values (all columns are complete).FALSEindicates that a row contains at least one missing value.
For example, using our sample salary_data data frame:
salary_data#> Name Age Salary
#> 1 Alice 25 50000
#> 2 Francisca NA 52000
#> 3 Fatima 30 NA
#> 4 David 35 55000complete.cases(salary_data)#> [1] TRUE FALSE FALSE TRUEThis indicates that:
Row 1 (
Alice): No missing values, so it is complete (TRUE).Row 2 (
Francisca): Missing value in theAgecolumn, so it is not complete (FALSE).Row 3 (
Fatima): Missing value in theSalarycolumn, so it is not complete (FALSE).Row 4 (
David): No missing values, so it is complete (TRUE).
5.8.2 Summarising Missing Data
After identifying that your dataset contains missing values, it’s essential to quantify them to understand the extent of the issue. Summarizing missing data helps you decide how to handle these gaps appropriately. To count the total number of missing values in your entire dataset, you can use the sum() function combined with is.na(). Remember the is.na() function returns a logical vector where each element is TRUE if the corresponding value in the dataset is NA, and FALSE otherwise. Summing this logical vector gives you the total count of missing values because TRUE is treated as 1 and FALSE as 0 in arithmetic operations.
Example:
Suppose you have a sampled airquality dataset:
airquality_data <- data.frame(
Ozone = c(41, 36, 12, 18, NA, 28, 23, 19, 8, NA),
Solar.R = c(190, 118, 149, 313, NA, NA, 299, 99, 19, 194),
Wind = c(7.4, 8, 12.6, 11.5, 14.3, 14.9, 8.6, 13.8, 20.1, 8.6),
Temp = c(67, 72, 74, 62, 56, 66, 65, 59, 61, 69),
Month = c(5, 5, 5, NA, NA, NA, 5, 5, 5, 5),
Day = c(1, 2, 3, 4, 5, 6, 7, 8, 9, 10)
)
airquality_data#> Ozone Solar.R Wind Temp Month Day
#> 1 41 190 7.4 67 5 1
#> 2 36 118 8.0 72 5 2
#> 3 12 149 12.6 74 5 3
#> 4 18 313 11.5 62 NA 4
#> 5 NA NA 14.3 56 NA 5
#> 6 28 NA 14.9 66 NA 6
#> 7 23 299 8.6 65 5 7
#> 8 19 99 13.8 59 5 8
#> 9 8 19 20.1 61 5 9
#> 10 NA 194 8.6 69 5 10To count the total number of missing values in this dataset, you would use:
There are 7 missing values in the entire data frame.
Missing Values Per Column:
This output indicates:
Ozonecolumn has 2 missing values.Solar.Rcolumn has 2 missing values.Windcolumn has 0 missing values.Tempcolumn has 0 missing values.Monthcolumn has 3 missing values.Daycolumn has 0 missing values.
For a column-wise summary, you can also use the inspect_na() function from the inspectdf package.
inspectdf::inspect_na(airquality_data)#> # A tibble: 6 × 3
#> col_name cnt pcnt
#> <chr> <int> <dbl>
#> 1 Month 3 30
#> 2 Ozone 2 20
#> 3 Solar.R 2 20
#> 4 Wind 0 0
#> 5 Temp 0 0
#> 6 Day 0 05.8.3 Strategies for Dealing with Missing Data
Managing missing data is a critical step in data preprocessing. There are several strategies available, depending on the nature of the data and the goals of the analysis.
5.8.3.1 Remove Missing Values
You can remove rows with missing values using na.omit() function:
cleaned_data <- na.omit(airquality_data)
cleaned_data#> Ozone Solar.R Wind Temp Month Day
#> 1 41 190 7.4 67 5 1
#> 2 36 118 8.0 72 5 2
#> 3 12 149 12.6 74 5 3
#> 7 23 299 8.6 65 5 7
#> 8 19 99 13.8 59 5 8
#> 9 8 19 20.1 61 5 9This method is simple and effective when the proportion of missing data is small. However, it can result in a significant loss of data if many rows contain missing values.
5.8.3.2 Replace Missing Values
Replace missing values with a default value using replace_na() from the tidyr package:
library(tidyverse) # tidyr is part of tidyverse
dataset <- airquality_data %>%
replace_na(list(Ozone = 17, Month = 3))
dataset#> Ozone Solar.R Wind Temp Month Day
#> 1 41 190 7.4 67 5 1
#> 2 36 118 8.0 72 5 2
#> 3 12 149 12.6 74 5 3
#> 4 18 313 11.5 62 3 4
#> 5 17 NA 14.3 56 3 5
#> 6 28 NA 14.9 66 3 6
#> 7 23 299 8.6 65 5 7
#> 8 19 99 13.8 59 5 8
#> 9 8 19 20.1 61 5 9
#> 10 17 194 8.6 69 5 10This approach ensures that missing values are replaced consistently, allowing for meaningful analysis without introducing bias.
5.8.3.3 Impute Missing Values
Missing values can be replaced with statistical measures such as the mean, median, or mode. For example, missing values in the Ozone column can be replaced with the mean, while missing values in the Month column can be replaced with the median:
library(dplyr)
airquality_data <- airquality_data %>%
mutate(
Ozone = ifelse(is.na(Ozone), mean(Ozone, na.rm = TRUE), Ozone),
Month = ifelse(is.na(Month), median(Month, na.rm = TRUE), Month)
)
# View the resulting dataset
airquality_data#> Ozone Solar.R Wind Temp Month Day
#> 1 41.000 190 7.4 67 5 1
#> 2 36.000 118 8.0 72 5 2
#> 3 12.000 149 12.6 74 5 3
#> 4 18.000 313 11.5 62 5 4
#> 5 23.125 NA 14.3 56 5 5
#> 6 28.000 NA 14.9 66 5 6
#> 7 23.000 299 8.6 65 5 7
#> 8 19.000 99 13.8 59 5 8
#> 9 8.000 19 20.1 61 5 9
#> 10 23.125 194 8.6 69 5 10If you don’t want to use ifelse, you can achieve the same result using the coalesce() function from dplyr. coalesce() replaces NA values by providing a fallback value.
library(dplyr)
airquality_data <- airquality_data %>%
mutate(
Ozone = coalesce(Ozone, mean(Ozone, na.rm = TRUE)),
Month = coalesce(Month, median(Month, na.rm = TRUE))
)
# View the resulting dataset
airquality_data#> Ozone Solar.R Wind Temp Month Day
#> 1 41.000 190 7.4 67 5 1
#> 2 36.000 118 8.0 72 5 2
#> 3 12.000 149 12.6 74 5 3
#> 4 18.000 313 11.5 62 5 4
#> 5 23.125 NA 14.3 56 5 5
#> 6 28.000 NA 14.9 66 5 6
#> 7 23.000 299 8.6 65 5 7
#> 8 19.000 99 13.8 59 5 8
#> 9 8.000 19 20.1 61 5 9
#> 10 23.125 194 8.6 69 5 10coalesce()is a simpler and more concise alternative when dealing with missing values.coalesce(Ozone, mean(Ozone, na.rm = TRUE))replacesNAvalues inOzonewith the computed mean.coalesce(Month, median(Month, na.rm = TRUE))does the same forMonthusing the median.
For more advanced imputation methods, you can use specialised packages like mice or Hmisc. Additionally, the bulkreadr package simplifies the process with the fill_missing_values() function:
-
Impute Specific Columns:
library(bulkreadr) fill_missing_values(airquality_data, selected_variables = c("Ozone", "Solar.R"), method = "mean" )#> Ozone Solar.R Wind Temp Month Day #> 1 41.000 190.000 7.4 67 5 1 #> 2 36.000 118.000 8.0 72 5 2 #> 3 12.000 149.000 12.6 74 5 3 #> 4 18.000 313.000 11.5 62 5 4 #> 5 23.125 172.625 14.3 56 5 5 #> 6 28.000 172.625 14.9 66 5 6 #> 7 23.000 299.000 8.6 65 5 7 #> 8 19.000 99.000 13.8 59 5 8 #> 9 8.000 19.000 20.1 61 5 9 #> 10 23.125 194.000 8.6 69 5 10 -
Impute All Columns in the Data Frame:
fill_missing_values(airquality_data, method = "median")#> Ozone Solar.R Wind Temp Month Day #> 1 41.000 190.0 7.4 67 5 1 #> 2 36.000 118.0 8.0 72 5 2 #> 3 12.000 149.0 12.6 74 5 3 #> 4 18.000 313.0 11.5 62 5 4 #> 5 23.125 169.5 14.3 56 5 5 #> 6 28.000 169.5 14.9 66 5 6 #> 7 23.000 299.0 8.6 65 5 7 #> 8 19.000 99.0 13.8 59 5 8 #> 9 8.000 19.0 20.1 61 5 9 #> 10 23.125 194.0 8.6 69 5 10
This approach helps to preserve the structure of the dataset and minimises data loss. However, it can introduce bias if the chosen statistic (e.g., mean or median) does not accurately represent the underlying data.
5.8.3.4 Flag Missing Data
You can create a new column to flag rows with missing values:
airquality_data %>% mutate(missing_flag = !complete.cases(.))#> Ozone Solar.R Wind Temp Month Day missing_flag
#> 1 41.000 190 7.4 67 5 1 FALSE
#> 2 36.000 118 8.0 72 5 2 FALSE
#> 3 12.000 149 12.6 74 5 3 FALSE
#> 4 18.000 313 11.5 62 5 4 FALSE
#> 5 23.125 NA 14.3 56 5 5 TRUE
#> 6 28.000 NA 14.9 66 5 6 TRUE
#> 7 23.000 299 8.6 65 5 7 FALSE
#> 8 19.000 99 13.8 59 5 8 FALSE
#> 9 8.000 19 20.1 61 5 9 FALSE
#> 10 23.125 194 8.6 69 5 10 FALSEUsing !complete.cases(.) ensures the missing_flag column accurately identifies rows with missing values (TRUE) while marking rows without missing values as FALSE. This binary flag is useful for quickly filtering or inspecting incomplete data.
Alternatively, instead of a binary flag, you can add a column that shows the number of missing values in each row:
#> Ozone Solar.R Wind Temp Month Day missing_count
#> 1 41.000 190 7.4 67 5 1 0
#> 2 36.000 118 8.0 72 5 2 0
#> 3 12.000 149 12.6 74 5 3 0
#> 4 18.000 313 11.5 62 5 4 0
#> 5 23.125 NA 14.3 56 5 5 1
#> 6 28.000 NA 14.9 66 5 6 1
#> 7 23.000 299 8.6 65 5 7 0
#> 8 19.000 99 13.8 59 5 8 0
#> 9 8.000 19 20.1 61 5 9 0
#> 10 23.125 194 8.6 69 5 10 0Adding a column like missing_count provides a numeric indicator of the total number of missing values in each row. This approach is particularly helpful when you need to assess the extent of missingness across the dataset or prioritise rows for further investigation
Consider the potential biases introduced by removing all rows with missing values. In which scenarios would you prefer imputation over removal?
5.8.4 Practice Quiz 5.3
Question 1:
Which function in R checks if there are any missing values in an object?
Question 2:
Which approach removes any rows containing NA values?
Question 3:
If you decide to impute missing values in a column using the median, what is one potential advantage of using the median rather than the mean?
- The median is always easier to compute.
- The median is more affected by outliers than the mean.
- The median is less influenced by extreme values and may provide a more robust estimate.
- The median will always be exactly halfway between the min and max values.
Question 4:
How would you replace all NA values in character columns with "Unknown"?
mutate(across(where(is.character), ~ replace_na(., "Unknown")))mutate_all(~ replace_na(., "Unknown"))Question 5:
What does the anyNA() function return?
- The number of missing values in an object.
-
TRUEif there are any missing values in the object; otherwise,FALSE.
- A logical vector of missing values in each row.
- A subset of the data frame without missing values.
Question 6:
You want to create a new column in a data frame that flags rows with missing values as TRUE. Which code achieves this?
df$new_col <- !complete.cases(df)-
df$new_col <- complete.cases(df)
-
df$new_col <- anyNA(df)
df$new_col <- is.na(df)
Question 7:
Before removing rows with missing values, what is an important consideration?
- Whether the missing values are randomly distributed across the data.
- Whether the dataset is stored in a data frame.
- Whether missing values exist in every column.
- Whether the missing values are encoded as
NA.
Question 8:
Why should the proportion of missing data in a row or column be considered before removing it?
- Removing rows or columns with minimal missing values may lead to excessive data loss.
- Columns with missing values cannot be visualized.
- Rows with missing values are always irrelevant.
- Rows with missing values should never be analyzed.
Question 9:
If a dataset has 50% missing values in a column, what is a common approach to handle this situation?
- Replace missing values with the column mean.
- Remove the column entirely.
- Replace missing values with zeros.
- Leave the missing values as they are.
Question 10:
What does the following Tidyverse-style code do?
library(dplyr)
airquality_data <- airquality_data %>%
mutate(Ozone = if_else(is.na(Ozone), mean(Ozone, na.rm = TRUE), Ozone))- Removes rows where
Ozoneis missing.
- Replaces missing values in
Ozonewith the mean of the column.
- Flags rows where
Ozoneis missing.
- Deletes the
Ozonecolumn if it has missing values.
5.8.5 Exercise 5.3.1: Handling Missing Data in the Television Company Dataset
This exercise will test your data cleaning skills using the data-tv-company.csv dataset, located in the r-data directory. If you do not already have the file, you can download it from Google Drive.
Dataset Metadata
This dataset was collected by a small television company seeking to understand the factors that influence how viewers rate the company. It includes viewer ratings and related measures. The variables in the dataset are as follows:
regard: Viewer rating of the television company (higher ratings indicate greater regard).
gender: The gender with which the viewer identifies.
views: The number of views.
online: The number of times bonus online material was accessed.
library: The number of times the online library was browsed.
Show1 to Show4: Scores for four different shows.
Tasks
-
Importing and Inspecting Data
- Locate the
data-tv-company.csvfile in ther-datadirectory. - Import the data into your R environment.
- Inspect the dataset for any missing values.
- Locate the
-
Strategies for Dealing with Missing Data
- Demonstrate at least four different methods for handling missing data in this dataset.
- Apply these methods to the imported data.
- Evaluate the methods and select the best approach based on your analysis.
5.9 Reflective Summary
In Lab 5, you developed essential skills in data transformation with R, including::
The Pipe Operator
|>: You learned to link functions in a logical sequence, enhancing code readability.Data Manipulation with
dplyr: You used core verbs—select(),filter(),mutate(),arrange(), andsummarise()—to reshape and refine your data.Summarisation and Grouping: By using
group_by()andsummarise(), you aggregated data to uncover patterns and derive insights.Handling Missing Data: You learned to detect and manage missing values, ensuring the quality of your analysis.
These techniques form a crucial step in the data analysis pipeline, enabling you to approach complex datasets with confidence and produce meaningful insights.
In the next lab, we will explore tidy data and joins. You will learn to reshape datasets and merge diverse data sources, converting raw data into structured, analysis-ready formats. This will pave the way for deeper insights and more efficient workflows.
The
|>operator (called a pipe) means “and then.” It passes the result of one function to the next.↩︎If you haven’t installed it yet, you can do so with
install.packages("palmerpenguins")and load it usinglibrary(palmerpenguins).↩︎Horst AM, Hill AP, Gorman KB (2020). palmerpenguins: Palmer Archipelago (Antarctica) penguin data. R package version 0.1.0. https://allisonhorst.github.io/palmerpenguins/. doi: 10.5281/zenodo.3960218.↩︎
Siegel, J. M. (2005). Clues to the functions of mammalian sleep. Nature, 437(7063), 1264-1271.↩︎
Using the same example, the syntax for
across()with column positions would be as follows:msleep |> select(name, contains("sleep")) |> mutate(across(c(2, 3, 4), ~ .x * 60))↩︎The
between()function simplifies the code and improves readability by combining the range condition into a single statement.↩︎While
slice()selects rows based on their numeric position (e.g., the first or last few rows), without reordering the entire dataset, thearrange()function is used to reorder all rows in the dataset according to the values of one or more specified columns.↩︎