6 Tidy Data and Joins
6.1 Introduction
Welcome to Lab 6! In Lab 5, you explored advanced data transformation techniques—including the pipe operator, key dplyr functions, and methods for handling missing data. Now, we will move on to organising and transforming your data using tidy data principles.
Tidy data involves arranging your dataset so that each variable occupies its own column, each observation its own row, and each type of observational unit its own table. Although tidying data may require some upfront effort, this structure greatly simplifies subsequent analysis and improves efficiency. With the tidyverse tools at your disposal, you will spend less time cleaning data and more time uncovering insights.
If you have ever struggled with reshaping datasets, merging data from multiple sources, or applying complex transformations, this lab is for you. The skills you acquire here are essential for real-world data analysis and will significantly enhance your proficiency in R.
6.2 Learning Objectives
By the end of this lab, you will be able to:
Understand Tidying Data:
Comprehend what tidy data is and why it is crucial for effective analysis.Reshape Data:
Convert datasets between wide and long formats using functions likepivot_longer()andpivot_wider()to prepare your data for analysis and visualisation.Separate and Unite Columns:
Useseparate()to split columns andunite()to combine them, thereby improving the structure and usability of your dataset.Combine Datasets Effectively:
Master various join operations indplyrto merge datasets seamlessly, resolving issues such as mismatched keys and duplicates.
By completing this lab, you will be equipped to transform messy, real-world datasets into analysis-ready formats, paving the way for insightful visualisations, robust models, and confident data analysis.
6.3 Prerequisites
Before you begin this lab, you should have:
Completed Lab 5 or have a basic understanding of data manipulation.
A working knowledge of R’s data structures.
Familiarity with the tidyverse packages, particularly dplyr.
6.4 The Principles of Tidy Data
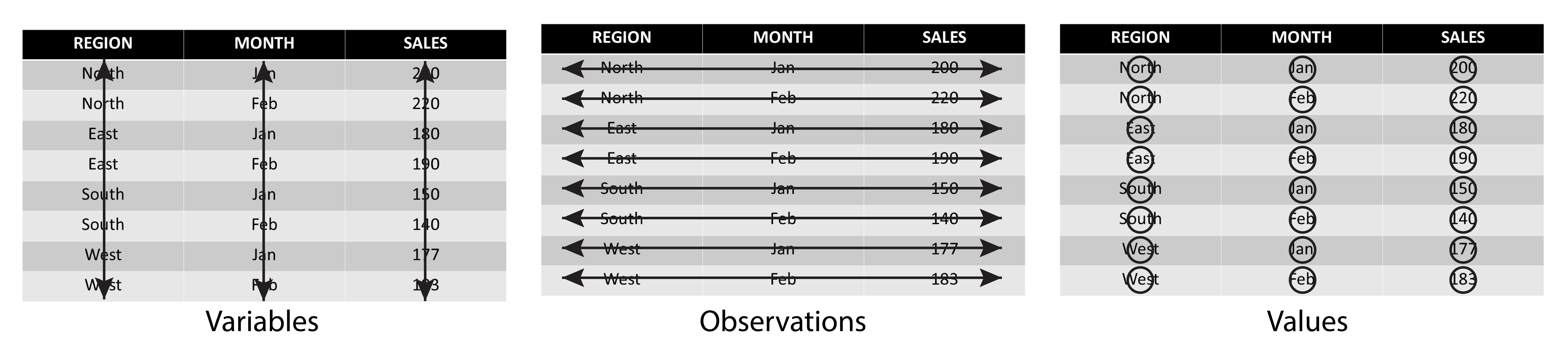
Tidy data is all about maintaining a clear and consistent structure. It is organised in such a way that:
Each variable forms a column.
Each observation forms a row.
Each cell contains a single value.
As illustrated in Figure 6.1, this structure makes it much easier to analyse and visualise data. Tools like tidyr and dplyr work best with data that adheres to these principles, minimising errors and streamlining the analysis process. In essence, tidy data provides the foundation for efficient data manipulation and reproducible research.
6.5 Experiment 6.1: Reshaping Data with tidyr
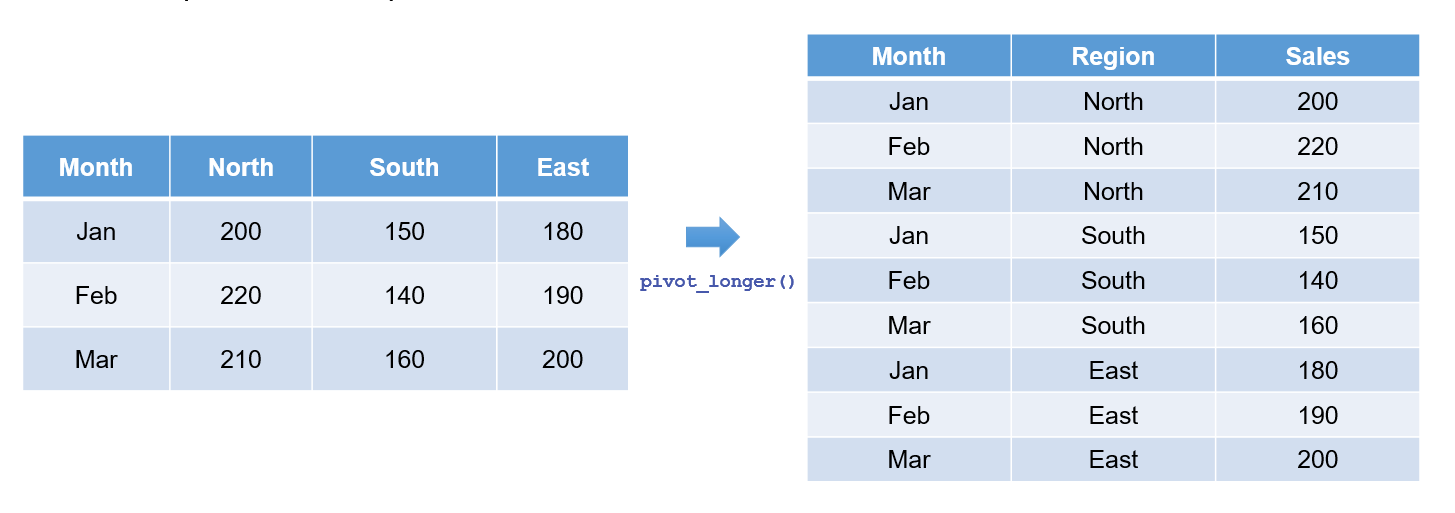
Since most real-world datasets are not tidy, they are often collected in a wide format, which can complicate detailed analysis. For instance, consider a sales manager who records monthly sales figures for each region in separate columns. Table 6.1 illustrates this wide-format layout:
| Month | North | East | South | West |
|---|---|---|---|---|
| Jan | 200 | 180 | 150 | 177 |
| Feb | 220 | 190 | 140 | 183 |
| Mar | 210 | 200 | 160 | 190 |
For many analyses—such as trend identification and visualisation—it is advantageous to reshape this data into a long format where each row represents a unique combination of region and month. Table 6.2 shows the transformed data:
| Region | Month | Sales |
|---|---|---|
| North | Jan | 200 |
| North | Feb | 220 |
| North | Mar | 210 |
| East | Jan | 180 |
| East | Feb | 190 |
| East | Mar | 200 |
| South | Jan | 150 |
| South | Feb | 140 |
| South | Mar | 160 |
| West | Jan | 177 |
| West | Feb | 183 |
| West | Mar | 190 |
This long format is particularly useful for time series analysis and visualisation, as it consolidates all pertinent information into common columns. The tidyr package, a core component of the tidyverse, provides efficient functions to perform these transformations.
6.5.1 Reshaping Data from Wide to Long Using pivot_longer()
The pivot_longer() function gathers multiple columns into two key-value columns. This process is essential when columns represent variables that you wish to convert into rows. Figure 6.2 illustrates this transformation:
pivot_longer()
The general syntax of pivot_longer() is:
pivot_longer(
cols = <columns to reshape>,
names_to = <column for variable names>,
values_to = <column for values>
)Where:
cols: A tidy-select expression specifying the columns to pivot into longer format.names_to: A character vector indicating the name of the new column (or columns) that will store the original column names incols.values_to: A string specifying the name of the new column that will store the corresponding values.
Example: Converting a Wide Dataset to Long Format
In the following example, we will convert the data in Table 6.1 into a long format using pivot_longer():
#> # A tibble: 3 × 5
#> Month North East South West
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Jan 200 180 150 177
#> 2 Feb 220 190 140 183
#> 3 Mar 210 200 160 190sales_data_long <- sales_data_wide |>
pivot_longer(
cols = c(North, East, South, West),
names_to = "Region",
values_to = "Sales"
)
sales_data_long#> # A tibble: 12 × 3
#> Month Region Sales
#> <chr> <chr> <dbl>
#> 1 Jan North 200
#> 2 Jan East 180
#> 3 Jan South 150
#> 4 Jan West 177
#> 5 Feb North 220
#> 6 Feb East 190
#> 7 Feb South 140
#> 8 Feb West 183
#> 9 Mar North 210
#> 10 Mar East 200
#> 11 Mar South 160
#> 12 Mar West 190The dataset is now prepared for further analysis, such as plotting sales figures by region or calculating averages.
6.5.2 Reshaping Data from Long to Wide Using pivot_wider()
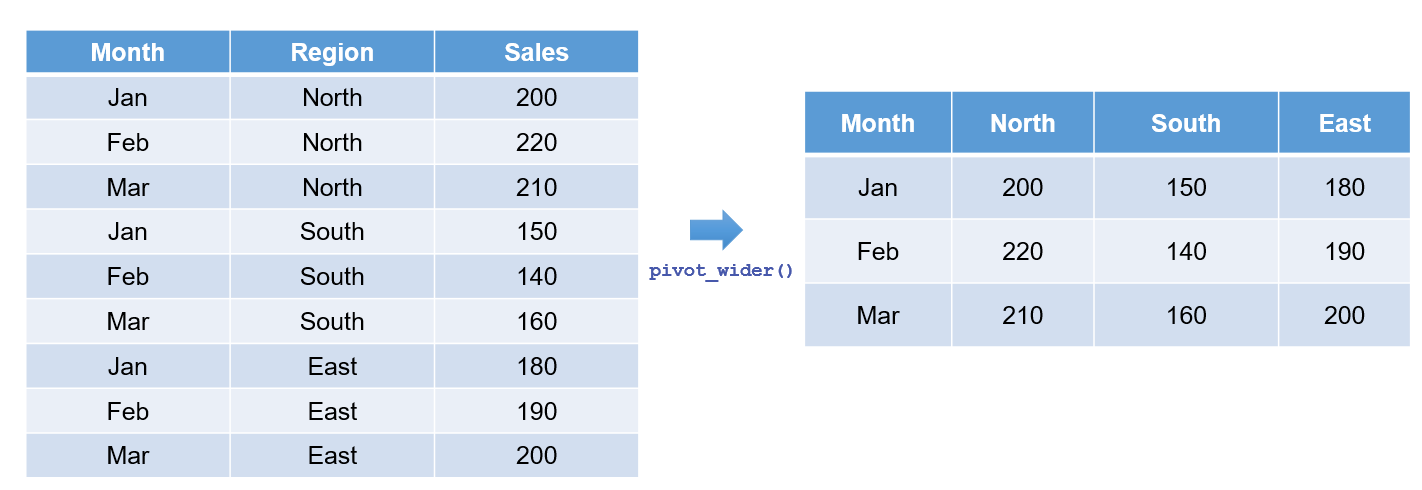
Conversely, the pivot_wider() function reverses the process by spreading key-value pairs into separate columns. This approach is particularly useful when you need to summarise data or create compact tables. Figure 6.3 demonstrates this transformation:
pivot_wider()
The general syntax of pivot_wider() is:
pivot_wider(
names_from = <column for new column names>,
values_from = <column for values>
)Where:
names_from: A tidy-select expression specifying the column(s) from which to derive the new column names.values_from: A tidy-select expression specifying the column(s) from which to retrieve the corresponding values.
Example: Converting a Long Dataset to Wide Format
Using the long-format sales data (sales_data_long), created earlier, we can convert it back to a wide format:
sales_data_long#> # A tibble: 12 × 3
#> Month Region Sales
#> <chr> <chr> <dbl>
#> 1 Jan North 200
#> 2 Jan East 180
#> 3 Jan South 150
#> 4 Jan West 177
#> 5 Feb North 220
#> 6 Feb East 190
#> 7 Feb South 140
#> 8 Feb West 183
#> 9 Mar North 210
#> 10 Mar East 200
#> 11 Mar South 160
#> 12 Mar West 190sales_data_wide <- sales_data_long |>
pivot_wider(
names_from = "Region",
values_from = "Sales"
)
sales_data_wide#> # A tibble: 3 × 5
#> Month North East South West
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Jan 200 180 150 177
#> 2 Feb 220 190 140 183
#> 3 Mar 210 200 160 190This transformation demonstrates how effortlessly data can be reshaped to meet the requirements of various analytical approaches.
6.5.3 Practice Quiz 6.1
Question 1:
Consider the following data frame:
sales_data_wide <- data.frame(
Month = c("Oct", "Nov", "Dec"),
North = c(180, 190, 200),
East = c(177, 183, 190),
South = c(150, 140, 160),
West = c(200, 220, 210)
)Which function would you use to convert this wide-format dataset into a long-format dataset?
-
pivot_long()
-
pivot_wider()
-
separate()
pivot_longer()
Question 2:
In the pivot_longer() function, if you want the original column names (“North”, “East”, “South”, “West”) to appear in a new column called “Region”, which argument would you use?
-
cols
-
names_to
-
values_to
names_prefix
Question 3:
Given the same data frame, which argument in pivot_longer() specifies the name of the new column that stores the sales figures?
-
names_to
-
values_to
-
cols
values_drop_na
Question 4:
What is the primary purpose of using pivot_wider()?
- To convert long-format data into wide format
- To combine two data frames
- To split a column into multiple columns
- To remove missing values
Question 5:
If you apply pivot_longer() on sales_data_wide without specifying cols, what is likely to happen?
- All columns will be pivoted, including the identifier column “Month”, leading to an undesired result.
- Only numeric columns will be pivoted.
- The function will automatically ignore non-numeric columns.
- An error will be thrown immediately.
Question 6:
Which package provides the functions pivot_longer() and pivot_wider()?
- dplyr
- tidyr
- ggplot2
- readr
Question 7:
The functions pivot_longer() and pivot_wider() are inverses of each other, allowing you to switch between wide and long formats easily.
- True
- False
Question 8:
In the following code snippet, what is the role of the cols = c(North, East, South, West) argument?
sales_data_long <- sales_data_wide |>
pivot_longer(
cols = c(North, East, South, West),
names_to = "Region",
values_to = "Sales"
)- It tells
pivot_longer()which columns to keep as they are.
- It specifies the columns to be pivoted from wide to long format.
- It defines the new column names for the output.
- It removes missing values from these columns.
Question 9:
After reshaping the data to long format, which of the following is a potential advantage?
- Easier to merge with other datasets
- Simplified time series analysis and visualisation
- Increased redundancy in the dataset
- Reduced number of observations
Question 10:
Which of the following best describes tidy data?
a) Each variable forms a column and each observation a row
b) Data is merged from multiple sources
c) Data is automatically plotted
d) Missing values are always removed
6.5.4 Exercise 6.1.1: Tidying the Pew Religion and Income Survey Data
This exercise tests your data tidying skills using the religion_income dataset, which can be found in the r-data directory. If you do not already have the file, you may download it from Google Drive.
Dataset Metadata
The Pew Research Trust’s 2014 survey compiled this dataset to examine the relationship between religious affiliation and income in the United States. It shows the proportions of sampled individuals from each religious tradition who fall into various income bands (e.g., <$10k, $10k-$30k, etc.). The dataset includes:
religion: The name of the religion.
Income Range Columns: Multiple columns corresponding to various income brackets (e.g.
<$10k,$10k-$30k, etc.). Each column represents the number of respondents falling within that income category. Some columns may also include descriptors such as “Don’t know/refused”.
Tasks
-
Importing and Inspecting Data
Locate the
religion_income.csvfile in ther-datadirectory.Import the data into your R environment.
Inspect the structure of the dataset and familiarise yourself with its variables.
-
Data Tidying
Reshape the dataset so that the various income range columns are gathered into two new variables: one for the income range (e.g.
income_range) and another for the corresponding number of respondents (e.g.count).Rename the resulting columns as necessary to ensure they are clear and descriptive.
Create a summary table that displays the total number of respondents per income range across all religions.
Produce a bar plot to visualise the distribution of respondents across the different income ranges.
Happy tidying!
6.6 Experiment 6.2: Splitting and Combining Columns
In this section, we will explore two essential functions from the tidyr package-separate() and unite()—that help restructure your data effectively.
6.6.1 Splitting Columns with separate()
The separate() function splits a single column into multiple columns. It offers several variations, each tailored to different splitting methods:
separate_wider_delim(): Splits a column using a specified delimiter.separate_wider_position(): Splits a column at fixed widths.separate_wider_regex(): Splits a column using regular expressions.
separate()
The general syntax is:
separate(data, col = <column to split>, into = <new columns>, sep = <separator>, remove = <logical flag>)Where:
data: The data frame to be transformed.col: The column to be split.into: A character vector specifying the names of the new columns.sep: A string or regular expression indicating where to split the column. If not specified, it defaults to splitting at non-alphanumeric characters.remove: A logical flag (defaultTRUE) indicating whether to remove the original column after splitting.convert: A logical flag (defaultFALSE); ifTRUE, it automatically converts the new columns to appropriate data types usingtype.convert().
Example 1:
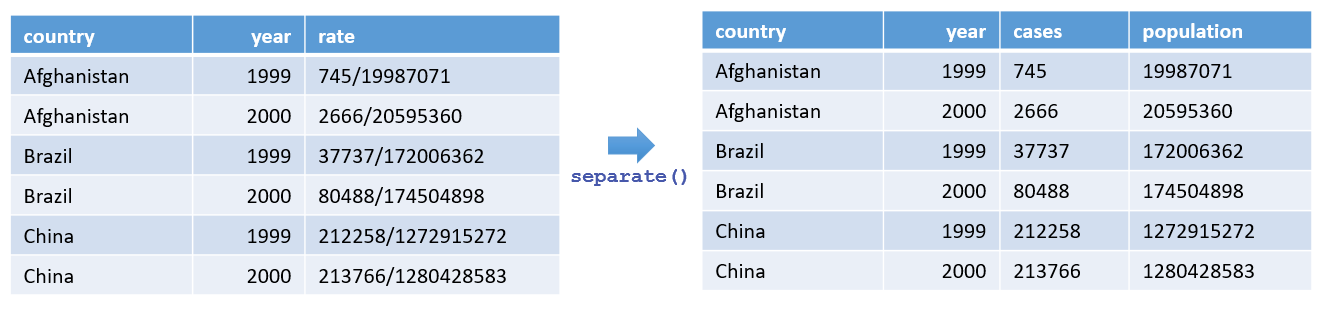
Below is a data frame showing tuberculosis (TB) rates in Afghanistan, Brazil, and China from 1999 to 2000. The "rate" column contains both cases and population values combined by a slash (/).
library(tidyverse)
# Create the data frame
tb_cases <- tibble(
country = c("Afghanistan", "Afghanistan", "Brazil", "Brazil", "China", "China"),
year = c(1999, 2000, 1999, 2000, 1999, 2000),
rate = c(
"745/19987071", "2666/20595360", "37737/172006362", "80488/174504898",
"212258/1272915272", "213766/1280428583"
)
)
tb_cases#> # A tibble: 6 × 3
#> country year rate
#> <chr> <dbl> <chr>
#> 1 Afghanistan 1999 745/19987071
#> 2 Afghanistan 2000 2666/20595360
#> 3 Brazil 1999 37737/172006362
#> 4 Brazil 2000 80488/174504898
#> 5 China 1999 212258/1272915272
#> 6 China 2000 213766/1280428583Since the "rate" column combines both cases and population, we can split it into two separate columns, "cases" and "population", using separate_wider_delim():
tb_cases |>
separate_wider_delim(cols = rate, names = c("cases", "population"), delim = "/")#> # A tibble: 6 × 4
#> country year cases population
#> <chr> <dbl> <chr> <chr>
#> 1 Afghanistan 1999 745 19987071
#> 2 Afghanistan 2000 2666 20595360
#> 3 Brazil 1999 37737 172006362
#> 4 Brazil 2000 80488 174504898
#> 5 China 1999 212258 1272915272
#> 6 China 2000 213766 1280428583This approach is particularly helpful when your data arrive in a combined format that needs to be split for meaningful analysis.
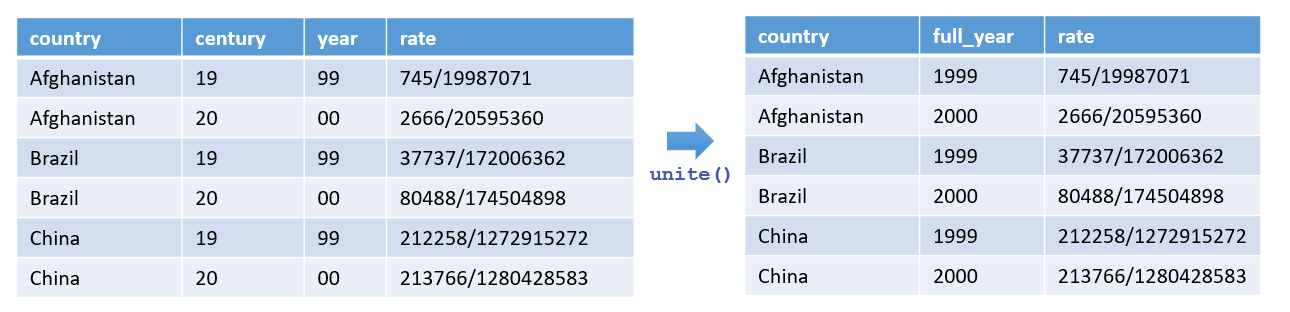
6.6.2 Combining Columns with unite()
Conversely, the unite() function merges multiple columns into one.
unite()
Its syntax is:
unite(data, col = <name of new column>, ..., sep = <separator>, remove = <logical flag>)Where:
data: The data frame to be transformed.col: The name of the new column that will contain the combined values....: The columns to combine.sep: The separator to insert between values (defaults to"_"if not specified).remove: A logical flag (defaultTRUE) indicating whether to remove the original columns after merging.
Example 2:
Imagine your TB dataset has separate "century" and "year" columns that you want to combine into a single "year" column:
# Create the data frame
tb_cases <- tibble(
country = c("Afghanistan", "Afghanistan", "Brazil", "Brazil", "China", "China"),
century = c("19", "20", "19", "20", "19", "20"),
year = c("99", "00", "99", "00", "99", "00"),
rate = c("745/19987071", "2666/20595360", "37737/172006362", "80488/174504898", "212258/1272915272", "213766/1280428583")
)
tb_cases#> # A tibble: 6 × 4
#> country century year rate
#> <chr> <chr> <chr> <chr>
#> 1 Afghanistan 19 99 745/19987071
#> 2 Afghanistan 20 00 2666/20595360
#> 3 Brazil 19 99 37737/172006362
#> 4 Brazil 20 00 80488/174504898
#> 5 China 19 99 212258/1272915272
#> 6 China 20 00 213766/1280428583You can merge "century" and "year" into a single column, also named "year", by specifying no separator:
tb_cases |> unite(year, century, year, sep = "")#> # A tibble: 6 × 3
#> country year rate
#> <chr> <chr> <chr>
#> 1 Afghanistan 1999 745/19987071
#> 2 Afghanistan 2000 2666/20595360
#> 3 Brazil 1999 37737/172006362
#> 4 Brazil 2000 80488/174504898
#> 5 China 1999 212258/1272915272
#> 6 China 2000 213766/1280428583This operation tidies your dataset by reducing two related columns into one, making future analysis more straightforward.
6.6.3 Practice Quiz 6.2
Question 1:
Given the following tibble:
tb_cases <- tibble(
country = c("Brazil", "Brazil", "China", "China"),
year = c(1999, 2000, 1999, 2000),
rate = c("37737/172006362", "80488/174504898", "212258/1272915272", "213766/1280428583")
)Which function would you use to split the "rate" column into two separate columns for cases and population?
Question 2:
Which argument in separate() allows automatic conversion of new columns to appropriate data types?
-
remove
autoconvertinto
Question 3:
Which function would you use to merge two columns into one, for example, combining separate “century” and “year” columns?
Question 4:
In the separate() function, what does the sep argument define?
- The new column names
- The delimiter at which to split the column
- The data frame to be merged
- The columns to remove
Question 5:
Consider the following data frame:
tb_cases <- tibble(
country = c("Afghanistan", "Brazil", "China"),
century = c("19", "19", "19"),
year = c("99", "99", "99")
)Which code correctly combines “century” and “year” into a single column “year” without any separator?
-
tb_cases |> unite(year, century, year, sep = "")
-
tb_cases |> separate(year, into = c("century", "year"), sep = "")
-
tb_cases |> unite(year, century, year, sep = "_")
tb_cases |> pivot_longer(cols = c(century, year))
Question 6:
When using separate(), how can you retain the original column after splitting it?
- Set
remove = FALSE
- Set
convert = TRUE
- Use
unite()instead
- Omit the
separgument
Question 7:
Which variant of separate() would you use to split a column at fixed character positions?
Question 8:
By default, the unite() function removes the original columns after combining them.
- True
- False
Question 9:
What is the main benefit of using separate() on a column that combines multiple data points (e.g. “745/19987071”)?
- It facilitates the conversion of string data into numeric data automatically.
- It simplifies further analysis by splitting combined information into distinct, analysable components.
- It merges the data with another dataset.
- It increases data redundancy.
Question 10:
Which argument in unite() determines the character inserted between values when combining columns?
-
separator
-
sep
-
col
delimiter
6.6.4 Exercise 6.2.1: Transforming the Television Company Dataset
This exercise tests your data cleaning skills using the television-company-data.csv dataset, which can be found in the r-data directory. If you do not already have the file, you may download it from Google Drive.
Dataset Metadata
This dataset was gathered by a small television company aiming to understand the factors influencing viewer ratings of the company. It contains viewer ratings and related metrics. The variables are as follows:
regard: Viewer rating of the television company (higher ratings indicate greater regard).gender: The gender with which the viewer identifies.views: The number of views.online: The number of times bonus online material was accessed.library: The number of times the online library was browsed.Shows: Scores for four different shows, each separated by a comma.
Tasks
-
Importing and Inspecting Data
Locate the
television-company-data.csvfile in ther-datadirectory.Import the data into your R environment.
-
Data Tidying
Create four new columns from the
Showscolumn, naming themShow1throughShow4.Create a new variable,
mean_show, calculated as the mean ofShow1toShow4.Examine how the mean show scores vary by
gender.
6.7 Experiment 6.3: Combining Datasets with Joins
When you’re working with real-world data, it’s common to have information scattered across multiple tables. In order to get a full picture and answer the questions you are interested in, you often need to merge these datasets. In this section, we will dive into how you can accomplish this in R using joins, a fundamental concept in both database management and data analysis.
6.7.1 The Role of Keys
Before you begin joining datasets, it’s crucial to understand what keys are and why they matter. In relational databases, a key is a column (or set of columns) that uniquely identifies each row in your dataset. When you join tables, keys determine how rows are matched across your datasets.
Primary Key: Think of this as the unique identifier for each record in a table. For example, in students table (Table 6.3 (a)), the
student_idis the primary key because it uniquely identifies each student.Foreign Key: This is a column in another table that links back to the primary key of the first table. In our case, the
student_idin an exam scores table (Table 6.3 (b)) would act as a foreign key, connecting scores to the corresponding students.
To illustrate, consider these two datasets in Table 6.3:
| student_id | name | age |
|---|---|---|
| 1 | Alice | 20 |
| 2 | Bob | 22 |
| 3 | Charlie | 21 |
| student_id | score |
|---|---|
| 1 | 85 |
| 2 | 90 |
| 4 | 78 |
In the Students table, the student_id column is the primary key that uniquely identifies each student. In the Exam Scores table, student_id is used as a foreign key to refer back to the Students table. We will use these two tables to illustrate the different types of joins.
6.7.2 Types of Joins
Joins are the backbone of relational data analysis, and the dplyr offers a family of join functions to merge tables, each with a specific way of handling matches and non-matches. They all follow a similar pattern:
join_function(x, y, by = "key_column")Where:
x: The first table (the “left” table).y: The second table (the “right table”).by: The column(s) to match on.
Let’s explore the main types: inner, left, right, and full joins. We wll use students and scores to see how each one works.
6.7.2.1 Inner Join
An inner join keeps only the rows where the key matches in both tables. If there’s no match, the row gets dropped. It’s the strictest join—think of it as finding the overlap between two circles in a Venn diagram.
The syntax is:
inner_join(x, y, by = "key_column")Example 1: Matching Students and Scores
Suppose you are a teacher who only wants to see data for students who are enrolled and took the exam. Using Tables 6.3 (a) and 6.3 (b):
students <- data.frame(student_id = 1:3, name = c("Alice", "Bob", "Charlie"), age = 20:22)
scores <- data.frame(student_id = c(1, 2, 4), score = c(85, 90, 78))
# Inner join
students |> inner_join(scores, by = "student_id")#> student_id name age score
#> 1 1 Alice 20 85
#> 2 2 Bob 21 90What’s Happening:
Alice (student_id 1) and Bob (student_id 2) appear because they’re in both tables.
Charlie (student_id 3) is missing—he didn’t take the exam, so there’s no match in scores.
Student_id 4 is also out—it’s in
scoresbut notstudents.
This join is perfect when you need complete data from both sides, like calculating averages for students with scores.
Think about how the result might change if we used left_join() instead?
6.7.2.2 The Left Join
A left join keeps every row from the left table (x) and brings along matching rows from the right table (y). If there’s no match, you get NA in the columns from y.
The syntax is:
left_join(x, y, by = "key_column")Example 2: Report Cards for All Students
Imagine you are preparing report cards and need every student listed, even if they missed the exam:
# Left join
students |> left_join(scores, by = "student_id")#> student_id name age score
#> 1 1 Alice 20 85
#> 2 2 Bob 21 90
#> 3 3 Charlie 22 NAWhat’s Happening:
All three students from students are here because it’s the left table.
Alice and Bob have their scores (85 and 90).
Charlie gets an NA for score—he’s enrolled but didn’t take the test.
Student_id 4 from scores is excluded—it’s not in the left table.
Use this when the left table is your priority, like ensuring every student gets a report card.
Consider what happens if the id column contains duplicates in either df1 or df2. This could result in more rows than expected in the joined dataset.
6.7.2.3 The Right Join
A right join flips it around: it keeps all rows from the right table (y) and matches them with the left table (x), filling in NA where needed.
The syntax is:
right_join(x, y, by = "key_column")Example 3: All Exam Scores
Now, suppose the exam office wants every score reported, even for students not in your class list:
# Right join
students |> right_join(scores, by = "student_id")#> student_id name age score
#> 1 1 Alice 20 85
#> 2 2 Bob 21 90
#> 3 4 <NA> NA 78What’s Happening:
All three scores from scores are included because it’s the right table.
Alice and Bob match up with their info from students.
Student_id 4 has a score (78) but no name or age (NA)—they’re not in students.
This is handy when the right table drives the analysis, like auditing all exam results.
6.7.2.4 The Full Join
A full join keeps everything—every row from both tables, matching where possible and using NA for gaps. It’s the most inclusive join.
The syntax is:
full_join(x, y, by = "key_column")Example 4: Complete Audit
For an audit, you want every student and every score, matched or not:
# Full join
students |> full_join(scores, by = "student_id")#> student_id name age score
#> 1 1 Alice 20 85
#> 2 2 Bob 21 90
#> 3 3 Charlie 22 NA
#> 4 4 <NA> NA 78What’s Happening:
Every student_id from both tables is here.
Alice and Bob are fully matched.
Charlie has no score (NA in score).
Student_id 4 has a score but no student info (NA in name and age).
This join is your go-to when you can’t afford to lose any data, like reconciling records.
6.7.3 Joins with Different Key Names
What happens when the keys don’t have the same name? Real datasets often throw this curveball—the primary key in one table might be club_code, while the foreign key in another is group_id. No worries—dplyr can still connect them.
Let’s try a new example:
Check them out:
clubs#> # A tibble: 3 × 2
#> club_code club_name
#> <chr> <chr>
#> 1 A01 Chess Club
#> 2 B02 Robotics Team
#> 3 C03 Art Societymembers#> # A tibble: 3 × 3
#> member_id name group_id
#> <int> <chr> <chr>
#> 1 101 Alice A01
#> 2 102 Bob B02
#> 3 103 Charlie D04Here, club_code is the primary key in clubs, and group_id is the foreign key in members. They’re different names, but their values (like “A01” and “B02”) match up.
Example:
You are a club coordinator and want to see who’s in which club, despite the naming mismatch. Use an inner join with a named vector in by:
clubs |> inner_join(members, by = c("club_code" = "group_id"))#> # A tibble: 2 × 4
#> club_code club_name member_id name
#> <chr> <chr> <int> <chr>
#> 1 A01 Chess Club 101 Alice
#> 2 B02 Robotics Team 102 BobWhat’s Happening:
The
by = c("club_code" = "group_id")tells R to matchclub_codefromclubswithgroup_idfrom members.Alice (A01) and Bob (B02) match their clubs.
Charlie (D04) is dropped—there’s no D04 in
clubs.
This trick works with any join type. For instance, a full join would include Charlie and the unmatched C03 club:
#> # A tibble: 4 × 4
#> club_code club_name member_id name
#> <chr> <chr> <int> <chr>
#> 1 A01 Chess Club 101 Alice
#> 2 B02 Robotics Team 102 Bob
#> 3 C03 Art Society NA <NA>
#> 4 D04 <NA> 103 CharlieJoins are your superpower for combining datasets in R. With inner_join(), you get the overlap; left_join() prioritizes the left table; right_join() favours the right; and full_join() keeps it all. Plus, you can handle differently named keys with a simple tweak to by.
6.7.4 Practice Quiz 6.3
Question 1:
Given the following data frames:
df1 <- data.frame(id = 1:4, name = c("Ezekiel", "Bob", "Samuel", "Diana"))
df2 <- data.frame(id = c(2, 3, 5), score = c(85, 90, 88))Which join would return only the rows with matching id values in both data frames?
a) left_join()
b) right_join()
c) inner_join()
d) full_join()
Question 2:
Using the same data frames, which join function retains all rows from df1 and fills unmatched rows with NA?
Question 3:
Which join function ensures that all rows from df2 are preserved, regardless of matches in df1?
Question 4:
What does a full join return when applied to df1 and df2?
- Only matching rows
- All rows from both data frames, with
NAfor unmatched entries
- Only rows from
df1
- Only rows from
df2
Question 5:
In a join operation, what is the purpose of the by argument?
- It specifies the common column(s) used to match rows between the data frames
- It orders the data frames
- It selects which rows to retain
- It converts keys to numeric values
Question 6:
If df1 contains duplicate values in the key column, what is a likely outcome of an inner join with df2?
- The joined data frame may contain more rows than either original data frame due to duplicate matches.
- The join will remove all duplicates automatically.
- The function will return an error.
- The duplicate rows will be merged into a single row.
Question 7:
An inner join returns all rows from both data frames, regardless of whether there is a match.
- True
- False
Question 8:
Consider the following alternative key columns:
df1 <- data.frame(studentID = 1:4, name = c("Alice", "Bob", "Charlie", "Diana"))
df2 <- data.frame(id = c(2, 3, 5), score = c(85, 90, 88))How can you join these two data frames when the key column names differ?
- Rename one column before joining.
- Use
by = c("studentID" = "id")in the join function. - Use an inner join without specifying keys.
- Convert the keys to factors.
Question 9:
What is a ‘foreign key’ in the context of joining datasets?
- A column in one table that uniquely identifies each row.
- A column in one table that refers to the primary key in another table.
- A column that has been split into multiple parts.
- A column that is combined using
unite().
Question 10:
Which join function would be most appropriate if you want a complete union of two datasets, preserving all rows from both?
6.7.5 Exercise 6.3.1: Relational Analysis with the NYC Flights 2013 Dataset
This exercise tests your relational analysis skills using the nycflights13 dataset, which is available as an R package. To access it, install and load the package.
Dataset Metadata
The nycflights13 dataset contains information on all 336,776 outbound flights from New York City airports (Newark Liberty International Airport (EWR), John F. Kennedy International Airport (JFK) and LaGuardia Airport (LGA)) in 2013, compiled from various sources, including the U.S. Bureau of Transportation Statistics. It is designed to explore the relationships between flight details, aircraft information, airport data, weather conditions and airline carriers. The dataset comprises five main tables:
flights: Details of each flight, including departure and arrival times, delays and identifiers such as
tailnum(aircraft tail number),origin(departure airport),dest(destination airport) andcarrier(airline code). This table contains 336,776 rows and 19 columns.planes: Information about aircraft, such as the manufacturer, model and year built, linked by the
tailnumkey. This table contains 3,322 rows and 9 columns.airports: Data on airports, including location and name, linked by the
faacode (used inflights$originandflights$dest). This table contains 1,458 rows and 8 columns.weather: Hourly weather data for New York City airports, linked by
originandtime_hour. This table contains 26,115 rows and 15 columns.airlines: Airline names and their carrier codes, linked by
carrier. This table contains 16 rows and 2 columns.
Tasks
-
Importing and Inspecting Data
Install and load the
nycflights13package in your R environment.Access the
flightsandplanestables, and inspect their structure to familiarise yourself with their variables.Identify the common key (
tailnum) that links flights and planes, and note any potential mismatches (for example, flights without corresponding plane data).
-
Relational Analysis with Joins
Perform an
inner_joinbetweenflightsandplanesusing thetailnumkey. How many rows are in the result, and why might this differ from the number of rows inflights?Perform a
left_joinbetweenflightsandplanesusing thetailnumkey. Compare the number of rows with the originalflightstable, and explain what happens to flights without matching plane data.Perform a
right_joinbetweenflightsandplanesusing thetailnumkey. How does this differ from theleft_joinresult, and what does it reveal about planes not used in flights?Perform a
full_joinbetweenflightsandplanesusing thetailnumkey. Describe how this result combines information from both tables, including cases with no matches.Create a summary table showing the number of flights per aircraft manufacturer (from
planes$manufacturer) after performing aleft_join. Handle missing values appropriately (for example, label flights with no plane data as “Unknown”).Produce a bar plot to visualise the distribution of flights across the top five aircraft manufacturers based on your summary table.
Happy joining!
6.8 Reflective Summary
In Lab 6, you have acquired advanced data transformation skills essential for effective data analysis:
Reshaping Data: You learned to convert datasets between wide and long formats using
pivot_longer()andpivot_wider(). Mastering these techniques is foundational for conducting time series analyses and creating visualisations.Separating and Uniting Columns: You explored how to split a single column into multiple columns with
separate()and combine several columns into one withunite(), thereby enhancing the structure of your data.Combining Datasets: You became familiar with various join operations in dplyr, enabling you to merge datasets seamlessly and manage issues such as mismatched keys and duplicates.
In the next lab, we will delve into data visualisation where will transform raw data into a visual language that reveals patterns, highlights trends, and conveys stories in ways that are easy to understand and interpret.